Message from RCSB PDB
PDB Reaches a New Milestone:
150,000+ Structures
With the March 20, 2019 weekly update, the PDB archive contained a record 150,145 structures.
Established in 1971, this central, public archive has reached this critical milestone thanks to the efforts of structural biologists throughout the world who contribute their experimentally-determined protein and nucleic acid structure data.
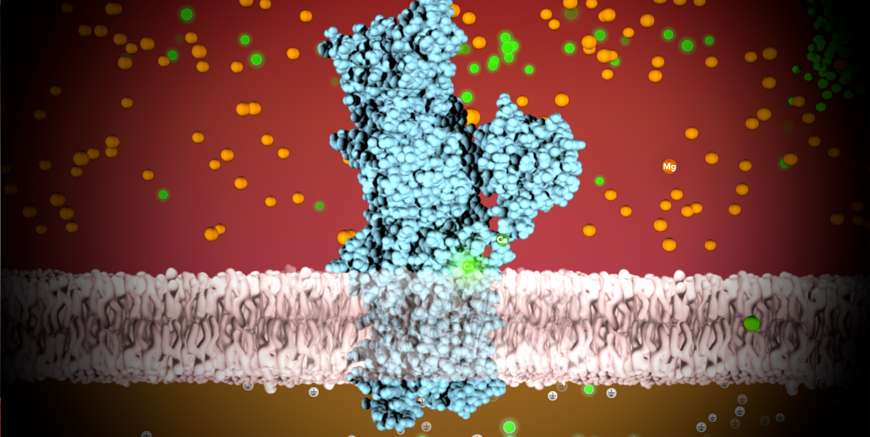
Four wwPDB data centers support online access to three-dimensional structures of biological macromolecules that help researchers understand many facets of biomedicine, agriculture, and ecology, from protein synthesis to health and disease to biological energy. The archive is large, containing more than 1.9 million files related to these PDB entries and requiring more than 512 Gbytes of storage.
The archive reached the landmark of 100,000 entries in 2014, the International Year of Crystallography. Since that record was set, the wwPDB organization achieved a significant milestone: the launch of a common global system for deposition, validation, and biocuration of PDB data for all supported experimental methods. Developed by the wwPDB team, the OneDep system unified deposition, annotation, and validation practices across all wwPDB partner sites.
OneDep also improved the availability of validation reports for both depositors (for review during biocuration) and data users (upon release). Building on expertise developed validating macromolecular crystallography, these reports assess the quality of each structure and highlight specific concerns, by examining the coordinates of the atomic model derived from either NMR and 3DEM, and by comparing the coordinates of the atomic model derived from NMR with primary experimental data therefrom. Readily interpretable summary information compares the quality of the atomic model with structural models from across the entire archive, thereby helping users of PDB data critically evaluate the quality of each archival entry.
More than 41,000 structures have been deposited, annotated, and validated using OneDep have been released into the PDB archive. OneDep uses the PDBx/mmCIF data format, which produces more uniform data, supports replacement of data files pre- and post-deposition, enhances communication with depositors, enables improved annotation, and provides validation reports based on recommendations from wwPDB expert task forces.
The scientific community eagerly awaits the next 150,000 structures and the invaluable knowledge these new data will bring. However, the increasing number, size and complexity of biological data being deposited in the PDB and the emergence of hybrid structure determination methods, which use a variety of biophysical, biochemical, and modelling techniques to determine the shapes of biologically relevant molecules, constitute major challenges for the management and representation of structural data. wwPDB will continue to work with the community to meet these challenges and ensure that the archive maintains the highest possible standards of quality, integrity, and consistency.
Many publications describe the development and future of the PDB archive and wwPDB organization, including Protein Data Bank (PDB): The Single Global Macromolecular Structure Archive (Methods in Molecular Biology, 2017), How community has shaped the Protein Data Bank (Structure, 2013), and Creating a Community Resource for Protein Science (Protein Science, 2012). A full list is available.

RCSB PDB wants to learn more about Molecule of the Month readers worldwide. Please take this brief survey and be entered into a drawing for a signed copy of The Machinery of Life by David Goodsell
| Snapshot: April 1, 2019 | |
|---|---|
| 150,393 | Released atomic coordinate entries |
| Molecule Type | |
| 139,500 | Proteins, peptides, and viruses |
| 3,316 | Nucleic acids |
| 7,547 | Protein/nucleic acid complexes |
| 30 | Other |
| Experimental Technique | |
| 134,425 | X-ray |
| 12,575 | NMR |
| 2,983 | Electron Microscopy |
| 275 | Hybrid |
| 135 | Other |
| Related Experimental Data Files | |
| 124,307 | Structure factors |
| 9,915 | NMR restraints |
| 3,667 | Chemical shifts |
| 3,042 | 3DEM map files |