DATA
QUERY, REPORTING AND ACCESS
Website Statistics
The RCSB PDB website at www.pdb.org began to utilize the
data from the wwPDB remediation project starting August 1, 2007.
Access statistics for this website are given below.
|
Month |
Unique Visitors |
Visits |
Bandwidth |
| Aug 2007 |
87494
|
225428
|
380.69
GB |
| Sep 2007
|
118631
|
294060
|
482.76
GB |
| Oct 2007
|
157581
|
389647 |
608.34
GB |
| Nov
2007 |
156243 |
373904 |
662.43
GB |
| Dec
2007 |
120351 |
284523 |
408.06
GB |
Automated
Downloads of PDB Data from ftp://ftp.wwpdb.org
As previously announced, the PDB archive has been moved to
ftp://ftp.wwpdb.org.
Updated weekly, this location maintains the files from the wwPDB
Remediation Project and all newly released files. The archive currently
contains approximately 350,000 files, including coordinate data
in PDB, mmCIF, and PDBML/XML formats, and experimental data. Since
the entire archive requires more than 70 GB of storage, fresh
downloads require a substantial amount of time. In December 2007,
more than 27 million files were downloaded from ftp.wwpdb.org.
During the same period, approximately 2.4 million files were downloaded
from the snapshot of unremediated data at ftp.rcsb.org. Users should
be aware that this site is no longer updated, and are strongly encouraged
to update any automatic scripts or bookmarks to
ftp://ftp.wwpdb.org.
Data files from the archive can be accessed online in a variety
of ways, including:
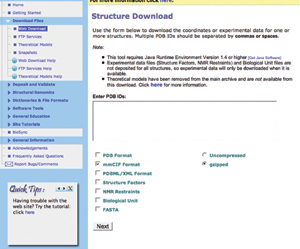
The online Structure Download tool
is accessible from the Download Files
section of the left hand menu. PDB IDs can be entered in the box
provided.
At ftp://ftp.wwpdb.org/pub/pdb/README,
users will find download information for downloading:
- A single file via ftp
- The entire archive via rsync
- All files in a given format (PDB, CIF, XML) via rsync
- The entire archive via ftp
- All files in a given format (PDB, CIF, XML) via ftp using tar
balls
RCSB
PDB Focus: Sorting Search Results
Following a search that produces multiple entries, the results
set can be sorted by choosing 'Sort Results' from the menu on the
left hand side of the page.
For most searches, the sorting options include: PDB ID, Release
Date, Residue Count, Resolution and Rank (useful with keyword searches).
An Advanced Search by sequence (Advanced Search >> Sequence
Features >> Sequence (Blast/Fasta)) allows the user to sort
results by PDB ID, formula weight and E value.
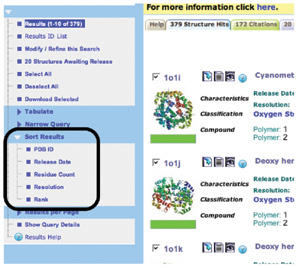
Structures matching your search
constraints can be sorted according to ID, release date,
residue count, resolution, and the rank of how closely they match
the query (shown in green).
Positions
Available at the RCSB PDB
The RCSB PDB has the following positions available:
- Senior Scientist/Scientific Software Developer (UCSD)
- Lead Web Architect (UCSD)
- Biochemical Information & Annotation Specialist (Rutgers)
For more information, select the
“Job Listings” link at
www.pdb.org.
|


