DATA
DEPOSITION AND ANNOTATION
ADIT 2.0 Offers Improved Deposition Process
 A new and improved version of ADIT, a tool for validating and submitting structures to the PDB archive, has been released.
ADIT 2.0 indicates file format errors and provides suggestions for solutions; automatically runs validation checks; and reviews the consistency between sequence and coordinates. Improved help information and documentation are provided. The forms for entering sequence information, structure title, author names, and citation information have also been streamlined.
This version of ADIT was developed based on user feedback and suggestions, and implemented after a period of testing. A new and improved version of ADIT, a tool for validating and submitting structures to the PDB archive, has been released.
ADIT 2.0 indicates file format errors and provides suggestions for solutions; automatically runs validation checks; and reviews the consistency between sequence and coordinates. Improved help information and documentation are provided. The forms for entering sequence information, structure title, author names, and citation information have also been streamlined.
This version of ADIT was developed based on user feedback and suggestions, and implemented after a period of testing.
ADIT and its related tools are available from deposit.rcsb.org. Deposition sessions currently in process will not be disrupted; depositors will not have to update any bookmarks or session ID.
Use Ligand Expo When Depositing Structures with Ligands
Ligand Expo (ligand-expo.rcsb.org) accesses chemical and structural information about all small molecule components found in PDB entries. It is based upon the Chemical Component Dictionary maintained by the wwPDB.
When depositing a structure with a ligand
- Search Ligand Expo for a chemical component that matches
your ligand
- If a match is found, use the corresponding three-character
code for the ligand in your coordinates
- If the ligand is not found, choose a new, unused three-
character code. When depositing this new ligand during
your ADIT deposition, upload the chemical name and
formula and/or a file showing the chemical image for the
new ligand into the Ligand Information section
Ligand Expo can search the Chemical Component Dictionary using uploaded data files (e.g., PDB, MOL/SDF, Refmac/ Phenix monomer library (mmCIF)); chemical name; formula; SMILES string; and ID code.
Searches for instances of ligands associated with macromolecular structures can also be performed at www.pdb.org using a variety of options, including the top bar Chemical Name/ID Search, Advanced Search, and the Chemical Structure Search. Users can then explore the ligand structures and the related PDB entries. Online screencasts are available at the RCSB PDB site to help users explore these features.
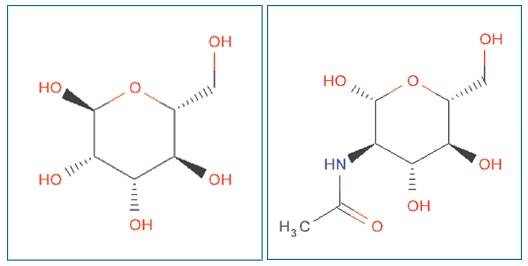
Ligand Expo can also be used to check and confirm the chemistry of a chemical component. Shown is the correct stereochemistry for alpha-D-mannose (ID: MAN) on the left, and for N-acetyl-D-glucosamine (ID: NAG) on the right.
Deposition Statistics
In the second quarter of 2010, 2312 experimentally-determined structures were deposited to the PDB archive. The entries were processed and annotated by wwPDB teams at the RCSB PDB, PDBe, and PDBj.
Of the structures deposited, 76.7% were deposited with a release status of "hold until publication"; 16.5% were released as soon as annotation of the entry was complete; and 6.8% were held until a particular date. 92.5% of these entries were determined by X-ray crystallographic methods; 6.8% were determined by NMR methods.
During the same time period, 1906 structures were released in the PDB, for a total of 4342 structures released in 2010.
wwPDB News from wwpdb.org
Validation Reports Available as PDFs
As part of the structure annotation process, wwPDB members provide depositors with detailed PDF reports that include the results of geometric and experimental data checking. These reports can be easily reviewed and shared by depositors.
As these validation reports provide an assessment of structure quality while keeping the coordinate data confidential, we encourage journal editors and referees to request them from depositors as part of the manuscript submission and review process. PDB validation reports are already required by the International Union of Crystallography (IUCr) journals1 as part of their submission process.
The reports are date-stamped, and display the wwPDB processing site logo. They contain essentially the same information, regardless of PDB annotation site. The reports will
continue to be developed and improved as we receive
recommendations from our Validation Task Forces for X-ray, NMR, EM, and small angle scattering methods, and as we
further develop our data deposition and processing procedures.
Questions about these reports and the annotation process may be sent to info@wwpdb.org.
Version 2 NMR Restraint Files Available from the wwPDB FTP
A new set of NMR restraint data files have been added to the wwPDB FTP with the June 30 update. These restraint files, identified as Version 2 files, are represented in NMR-STAR 3.1 format, contain current PDB atom nomenclature, and provide accurate atom-level correspondences to the NMR model coordinate files in the current archive. Restraint files containing restraint data as originally deposited (Version 1 files) will remain on the site and will continue to be updated regularly as new NMR entries are released.
The Version 2 NMR restraint files were generated for the wwPDB by the BMRB in collaboration with PDBe and the Centre for Molecular and Biomolecular Informatics/Institute for Molecules and Materials at the Radboud University Nijmegen. NMR restraints were parsed from their original format (Version 1), and harmonized with the coordinates using the software packages Wattos (BMRB; and CMBI/IMM), FormatConverter and NMRStarExport (PDBe), and the CCPN framework. The complexity of this process may have led to minor modifications or loss of data in the Version 2 restraint files due to parsing or conversion errors. The PDB coordinate file and the Version 1 restraint files remain the primary reference for these data. More information about the process used to generate these files is available.2-4
The initial release of the Version 2 NMR restraint files added more than 5700 new files (~310 Mbytes) to the FTP site. Version 2 restraint files for new PDB entries will be processed and made available after the PDB entry has been released. Version 1 restraint files for new PDB entries will continue to be released weekly.
NMR restraint files are named using extension .mr, as in "1abc.mr.gz" for PDB ID 1abc. Version 2 restraint files will be named "1abc_mr.str.gz", where "str" identifies the NMR-STAR (V3.1) data format. All restraint files are compressed (.gz) using the GNU gzip program.
Version 1 restraint files will remain in their current directory structure. Version 2 restraint files are stored in directories named by the middle two characters of their PDB ID in: ftp.wwpdb.org/pub/pdb/data/structures/divided/nmr_ restraints_v2/
These data files are linked symbolically to the directory: ftp.wwpdb.org/pub/pdb/data/structures/all/nmr_ restraints_v2/
|

 A new and improved version of ADIT, a tool for validating and submitting structures to the PDB archive, has been released.
A new and improved version of ADIT, a tool for validating and submitting structures to the PDB archive, has been released.

