Education Corner
 Takanori Nakane graduated from Kyoto University's School of Medicine in 2010. After passing Japan's national board exam and qualifying as a medical doctor, Nakane decided to enter a four-year PhD course in X-ray crystallography at Kyoto University instead of clinical practice. While working on "wet" experiments, he realized that he is more attracted to "dry" (computational) aspects of science.
Takanori Nakane graduated from Kyoto University's School of Medicine in 2010. After passing Japan's national board exam and qualifying as a medical doctor, Nakane decided to enter a four-year PhD course in X-ray crystallography at Kyoto University instead of clinical practice. While working on "wet" experiments, he realized that he is more attracted to "dry" (computational) aspects of science.Over the last year, Nakane has been developing molecular viewers for Web browsers and mobile devices, including the NDKMol molecular viewer that been incorporated with the RCSB PDB Mobile app. Nakane also contributes to several open-source projects, including PyMOL and Jmol.
Visit webglmol.sourceforge.jp to download Nakane's GLmol (Molecular viewer on WebGL/Javascript) and ESmol & NDKmol (Molecular viewer for Android).
Dr. Nakane may be reached at nakane.t@gmail.com.
As of December 2016, the RCSB mobile app will be retired. Please read the following news article.
Molecular Visualization, Online and On-the-Go
by Takanori Nakane M.D.
Molecules and Computers
I became fascinated by molecules starting in high school, when I found an old edition of Lehninger's Principles of Biochemistry on my father's bookshelf. I was quickly captivated by complex structures of biomolecules, and found the cyclization reaction leading from epoxysqualene to lanosterol especially impressive. In this reaction, four rings and eight stereocenters are created in a perfectly controlled manner. Ever since then, I have been deeply immersed in the world of chemistry.
Another enduring interest has been computer programming. When I was an eight year-old boy, my father gave me a very, very old computer and let me play with it freely. The CPU was 8-bit and had a main memory of only four kilobytes! Although it was not powerful at all, I had perfect control over the hardware. Since there wasn't an operating system or any device driver to be concerned about, I was able to read and write anything at any memory address and directly send commands to peripheral chips. It was an invaluable experience and taught me to make best of limited computational resources.
Making Structures More Accessible
When I was an undergraduate student, I noticed that biology students tended to learn compounds and enzymes only as names to be memorized, with little attention given to their structures. Similarly, only a few medical students look at the chemical structures of drugs and their target proteins. Even teachers tend to ignore molecular structures, and mention only their names. I believe these are missed opportunities to deepen one's understanding of molecules and to appreciate their beautiful structures. However, once teachers and students are introduced to the study of molecular structures and discover that they are interesting (RCSB PDB's Molecule of the Month is a good starting point), they too are able share my fascination with structure. My work is motivated by the idea that by increasing exposure, more people will become familiar with structures. What if more websites showed molecules in an interactive manner?
Another motivation for developing mobile tools is quite personal. Every time I come across a new compound, I search for the structural formula and copy it to my notebook by hand to get familiar with it. With a new protein, I check the PDB to see if its structure is determined. But I am not always in front of my computer!
When you are with your own computer, you can use any software you have. But what if you want to show a structure to your friend or colleague on another machine and you may be unable to install a new program? You need a molecular viewer that works within web browsers. Also, when you are listening to a lecture or a seminar, you might not be near a computer. However, since most of us carry smart phones or tablets with us all day long that use web browsers, I thought that developing a molecular viewer for the web would cover all situations. So my work on GLmol began.
GLmol: Molecular viewer for browsers
My work on GLmol, a molecular viewer for web browsers, began about a year ago. It is written in JavaScript alone, requiring no other plugins such as Flash or Java.
Most websites show structures of compounds and proteins only as static images. Some sites, including the RCSB PDB, use Java-based molecular viewers such as Jmol. While they are relatively fast once launched successfully, loading structures can be very slow because browsers have to first load the Java runtime environment. Occasionally browsers can be unresponsive during initialization. So I wanted to develop a molecular viewer that does not rely on anything but the web browser itself.
Fortunately, recent browser development has achieved substantial improvement in the speed of Javascript engines. Furthermore, a new enhancement to HTML called WebGL (Web Graphics Library) enables web pages to harness hardware acceleration for 3D rendering. GLmol is based on these technologies.
Pursuing Performance
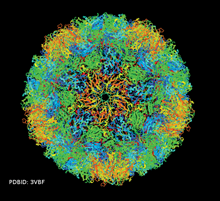
GLmol can handle very large molecules such as the enterovirus of PDB ID 3VBF. webglmol.sourceforge.jp/
glmol/viewer.html?pdb:3vbf (enable "biological assembly"in "View" tab)
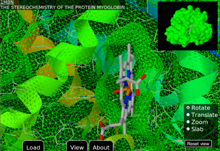
GLmol can even calculate molecular surfaces. See webglmol.sourceforge.jp/
surface.html for details.
Shown: PDB ID 1mbn.
Twenty years ago, molecular visualization required expensive graphics workstations. Today, thanks to powerful processors found in desktop computers, real-time rendering of molecules no longer poses the same challenge. Mobile platforms, however, are an exception. While most computers have four to eight CPU cores running at more than two gigahertz, most tablets have only two cores running at one gigahertz. Their memory is at most one gigabyte. This corresponds to typical desktop computers ten years ago. Therefore, simply porting existing molecular visualization software designed for desktop computers to the mobile platform will not succeed. It potentially might work for relatively small proteins but rendering would be very slow for large molecules. Programs running on web browsers have another limitation even when running on a desktop computer. Since they are written in a scripting language, they run several times slower than native applications.
GLmol was written and designed from scratch. First, I wrote a PDB file reader and a very simple "sphere" renderer, which draws atoms as spheres. Then I implemented "line" and "stick" renderers for bonds. Although this was sufficient for small molecules, it was practically useless for large biological macromolecules. As proteins are made up of tens of thousands of atoms, drawing all bonds and atoms quickly fills up the entire screen and makes it visually incomprehensible. Therefore, using a more simplified "ribbon" representation was necessary. However, this is not a trivial endeavor. A ribbon representation traces the protein's backbone, or alpha carbons. To display it as a ribbon, I had to consider what width and thickness to use. The ribbons also had to be smooth; otherwise, beta sheets would wiggle up and down. Of course, there are well-established algorithms for ribbon rendering. But I could not use them without significant modification, because they were too complicated or required too much memory. After trying various approaches, I chose one that gave the best results within the limited capacity of Javascript and WebGL (Web Graphics Library).
The result: GLmol can smoothly display very large molecules, even ribosomes and viral capsids!
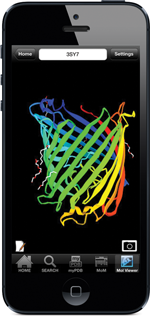
Screenshot of outer membrane carboxylate channel seen in PDB ID 3sy7 using the NDKmol viewer in RCSB PDB Mobile
Targeting Mobile Devices
At first I thought GLmol would cover desktop computers (Windows, Mac OS, Linux) and mobile devices (Android, iOS). However, I was disappointed to realize that WebGL was not yet available on most mobile devices. So I rewrote GLmol in C++, ported it to the Android OS platform, and called it NDKmol. I was pleased to find out that NDKmol works smoothly even on my old smart phone running Android 1.6. Then I ported NDKmol's core to iOS, which was adapted into the RCSB PDB Mobile App by Dr. Greg Quinn. NDKmol retains GLmol's very fluid interactivity, and can comfortably display very large molecules.
Biological Assemblies and Crystal Packing
A unique feature of my viewers is the capacity to display biological assemblies and crystal packing based on symmetry operations described in the PDB file. This functionality appears to be missing in most molecular rendering programs for mobile devices, which by default pull data from the coordinate file describing the asymmetric unit, rather than the coordinate file describing the complete biological assembly. This can be very confusing for people without training in structural biology. For example, students might want to look at the structure of potassium channel by the Nobel Prize winner Professor Roderick MacKinnon (PDB ID 1K4C). If they download the asymmetric unit file, they will never see the pore! Fortunately, my viewers use the biological assembly information to display the full structure. But they don't download the coordinate files for the complete biological assembly as that would consume too much memory. Instead, the viewers load the coordinate file describing the asymmetric unit alone, and let the GPU generate the correct symmetry. This process lets my viewers display large molecular assembly (like viral capsids) with ease.

Enabling the display of the biological assembly in GLmol for the potassium channel PDB ID 1K4C. webglmol.sourceforge.jp/glmol/
viewer.html?pdb:1K4C
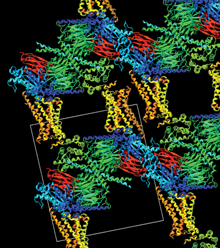
Crystal packing of adrenergic receptor-G protein complex (PDB ID: 3SN6). The receptor is in yellow, nanobody in red, G alpha in blue, G beta in green, and G gamma in cyan. webglmol.sourceforge.jp/
glmol/viewer.html?pdb:3sn6 (enable "crystal packing"in "View" tab)
Similar attention needs to be paid to the effects of crystal packing when examining structures determined by X-ray crystallography. Molecules touch each other in a crystal. This interaction might change the conformation of side chains and flexible loops. By applying crystallographic symmetry operations, my viewers can reconstruct how molecules are packed in the crystal. It's then apparent which part of the molecule might be potentially affected by packing interaction. This may sound very technical, but looking at features such as crystal packing is fun; for example, look at structure 3SN6 by this year's Nobel Prize winner Professor Brian Kobilka. Here the crystal is held together by dense layers of G proteins, nanobody and T4 lysozyme. All of these constituents were necessary to grow well-diffracting crystals of this amazing complex.
As an open source software
The source code for GLmol and NDKmol are made public under LGPL3 license. GLmol is also released under an MIT license that allows users to adapt the program in any way provided that the copyright notice is kept intact. Dozens of universities, research institutes, e-learning advocates and commercial companies have contacted me to integrate GLmol into their programs and to ask me for feature enhancements. This has been very encouraging and rewarding.
The source code is also very compact and simple. Actually, the entire code for GLmol is smaller than PyMOL's code for cartoon representation alone. The code is a good starting point to learn programming, how to parse PDB files, how to detect bonds, and how to build ribbons from atomic coordinates. You can also learn more general techniques such as OpenGL, HTML5 and multi-touch support. I learned a lot (and continue learning) by reading the source code of open source software and want to keep this cycle going. This is indeed what is happening now. A master course student built a sequence alignment viewer on GLmol. Several educators took GLmol and had summer students implement more features. This is again very stimulating for me.
I have enjoyed developing these molecular viewers, and hope that everyone has a nice time using them!
