| EDUCATION
CORNER

CHRISTOPHER SMITH is the Director of Education, Outreach & Training at the Center for Theoretical Biological Physics housed at the University of California San Diego (ctbp.ucsd.edu).
The NSF-funded CTBP, a collaboration between UCSD and The Salk Institute for Biological Studies, encompasses a broad array of research and training activities at the forefront of the biology-physics
interface. CTBP's mission is to conduct and foster research aimed at providing a quantitative framework for unraveling the workings of complex biological processes. Research within CTBP focuses on the following broadly defined areas: Cellular Tectonics Gene Regulatory Networks, and Computational Approaches to Intracellular and Intercelllular Communication.
Christopher's research interests encompass DNA and protein sequence and structural analysis and the computational tools facilitating these analyses. Current research focuses on the
development of user-friendly interfaces for computational tools and databases for the end-user (researcher and educator). He has developed web-based resources for educators (e.g., the CMSMBR, Discovery Tools), IT training workshops for educators, health-care professionals and researchers and is actively engaged in K-16 education projects. Christopher's education interests encompass the integration of information technology and science into K-16 and informal education.
The Anatomy of Vision:
Insights into Molecular Modeling
for the
Middle Schooler
by Christopher M. Smith, Ph.D
Most young scientists (middle/high schoolers) today are pretty savvy when it comes to the use of computers, cell phones, and social media networks. And they are very adept at using these tools for entertainment, so our challenge as educators is how to redirect this expertise and familiarity to further their education. As scientists, we can easily say that visualizing a protein in 3D using a molecular graphics program is pretty cool, but the reality is it pales in comparison to the 3D animations students use in computer gaming.1 So how do we engage these technologically savvy students? The answer (or at least one of them) lies in making the science topic relevant to them and in presenting it within the context of the world around them. A topic cannot be made relevant to them if it is taught in isolation from the rest of the world (or all other topics), so it must be integrated! This integration is the subject of this exercise to teach middle school (and sometimes sixth-grade) students molecular modeling; what it is, why it is important, and how it is used to advance our knowledge.
The end point of this exercise is to have the students use CN3D (or any other visualization application) to explore the various structural elements of the rhodopsin protein (1gu8). Keep in mind that my science objective is for the students to learn something about molecular modeling; everything else that I talk about is just a set of tools to engage their interest and active participation. I generally begin my molecular modeling presentation with a few comments about computer gaming (an interest of theirs), and drawing analogies to molecular modeling (an interest of mine). But the real story - the real fun begins when we start to talk about the anatomy and physiology of the eye. This is a topic that appears to always be interesting to the students, especially the visually impaired. You can use any general anatomical rendition of the eye that is available via practically any web resource. My preference is the very simple Human Eye image (Figure 1) because it depicts a cross-section of the eye with insets focusing on smaller and smaller details of that anatomy, i.e., general eye anatomy, the structure of the retinal tissue layer, a close-up of a light-sensing Rod, and a depiction of the rhodopsin protein in the Rod membrane. A description and walk-through of general eye anatomy includes exhaustive comments about protein composition and packing in the cornea to maintain clarity while providing strength and elasticity. I place an extreme emphasis on the health of the eye, especially the cornea, and perhaps scare the students a little with a couple of analogies and interesting discussions on the nature of proteins. For example, the cornea is much like the white of a chicken egg and it can and will turn irreversibly white if exposed to extremes of heat or chemicals (e.g., acetone found in nail polish remover - you can actually demonstrate the latter in class, which really catches their attention). I usually follow with a short, general description of protein folding and unfolding, and the concept of conformational changes. This is followed by an explanation and demonstration of how a conformational change in rhodopsin is elicited by a photon of light. This is vision in the making. The demonstration is a beautiful animation of the photoisomerization process as a photon of light hits a membrane embedded rhodopsin protein (see www.blackwellpublishing.com/matthews/rhodopsin.html). As students view the animation (again and again!), I also introduce the concept of light (photons), elements of protein secondary structure, and a description of retinal; where it is from and why it is important. For example, I usually ask the students if they have ever heard the expression, eat your carrots, they are good for you (your vision), then follow by introducing the nomenclature of organisms, e.g., the carrot (Daucus carota), and how its scientific name is related to the compound carotene, a metabolic precursor to retinal - the key ingredient of rhodopsin, essential for sight. Now that the students have grasped the relationships, i.e., these many topics are taught in context, we begin our exploration of the rhodopsin. You can start with Jmol, but many students like the stand-alone aspect and additional functionality of independent programs, so I invariably use Cn3D (NIH; www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). Once each student has Cn3D up and running with rhodopsin on the screen, you can walk through the various secondary structural elements, including the location of retinal, and talk about the significance of the parallel grouping of alpha helices, hydrophobicity, etc. The important thing is that the students are allowed to explore the model and ask questions. I also strongly emphasize that students try different visualization parameters (sticks, balls; color-schemes, etc.) and when they ask me What is it? I ask them what they see and how it is different from their previous representation. I interject into their answers additional detail on the structure that the particular parameter highlights. And soon enough, the students are exploring all the advanced, nook and cranny features of the program - not because I said so, but because of their curiosity. Sometimes the students will want to explore other proteins, so you may want to be prepared with a list of suggestions. My recommendations are protein toxins in the venom of cool animals; Charybdotoxin (Death Stalker Scorpion; 1tsk), Conotoxin (Cone Shell; 1hje), Tetrodotoxin (Fahaka Puffer Fish; 1rmk), and Convulxin (Rattlesnake; 1uos). The other interesting tidbit that can be added to the toxin structure discussion is the relative sizes of these proteins, and the significance this has physiologically and chemically as these toxins are distributed throughout the body (quickly) and with such drastic, immediate effects.
A final note - if you do this exercise with your students, be prepared to field questions for days and weeks later as the students do their own exploration of the Protein Data Bank.
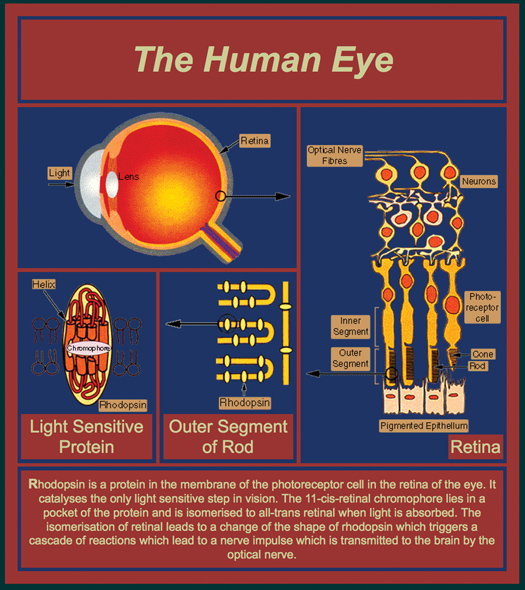
Figure 1.
Human Eye image available from www2.mrc-lmb.cam.ac.uk/groups/GS/eye.html (reprinted courtesy of Gebhard Schertler, Paul Scherrer Institute, Switzerland)
1. For an example, see Crystal structure of a monomeric retroviral protease solved by protein folding game players. F. Khatib, F. DiMaio, S. Cooper, M. Kazmierczyk, M. Gilski, S. Krzywda, H. Zabranska, I. Pichova, J. Thompson, Z. Popović, M. Jaskolski, D. Baker (2011) Nature Structural & Molecular Biology doi:10.1038/nsmb.2119 |


