RCSB PDB's website was visited each month by an average of 320872 unique visitors and 737293 unique visits. A total of 19479 GB of data were accessed.
| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2013 | 308327 | 703069 | 1785.92 GB |
| February 2013 | 317729 | 683177 | 1866.81 GB |
| March 2013 | 346548 | 765109 | 1529.32 GB |
| April 2013 | 369516 | 811545 | 1701.94 GB |
| May 2013 | 337920 | 765033 | 1669.93 GB |
| June 2013 | 291409 | 682857 | 1387.67 GB |
| July 2013 | 263698 | 655005 | 1392.55 GB |
| August 2013 | 259057 | 623656 | 1228.01 GB |
| September 2013 | 310025 | 733943 | 1505.25 GB |
| October 2013 | 383350 | 874037 | 1855.44 GB |
| November 2013 | 375120 | 864387 | 2000.50 GB |
| December 2013 | 287774 | 685699 | 1555.97 GB |
Web Services provide remote access to RCSB PDB resources to help software developers build tools that efficiently interact with PDB data. The RESTful API offers:
Two new tables provide access to drug and drug target information from DrugBank that are mapped to PDB entries with each weekly update. These tables, accessed from the Tools menu, can be searched by generic or brand drug name, filtered, and sorted.
The Drugs Bound to Primary Targets Table lists drugs bound to primary target(s), or a homolog of primary target(s), i.e., co-crystal structures of drugs.
From this view, click on the tab for Primary Drug Targets to view related structures in the PDB, regardless if the drug molecule is part of the PDB entry (e.g., apo forms of drug targets, drug target with different bound ligands). Biotherapeutics, such as complexes with monoclonal antibodies, are included.
A detailed description of these Drug and Drug Target Mapping tables is online.
A gene name option is available from the simple search bar at the top of RCSB PDB pages. To begin, start typing the name of a gene (for example brca) and a list of possible suggestions will appear:
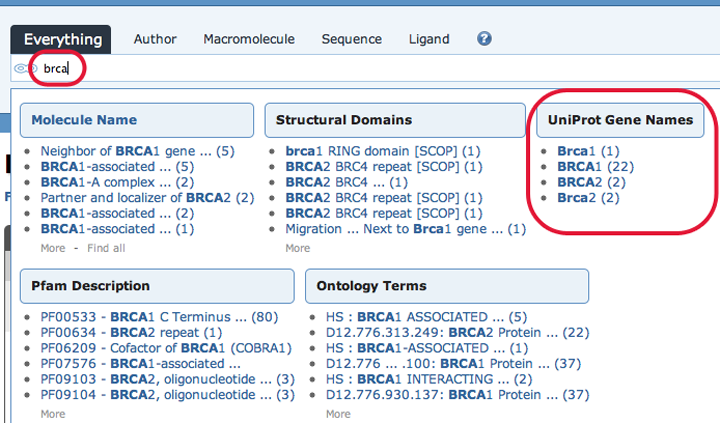
Then, select one of the suggestions in the "UniProt Gene Name" category. This will return a corresponding Protein Feature View page:
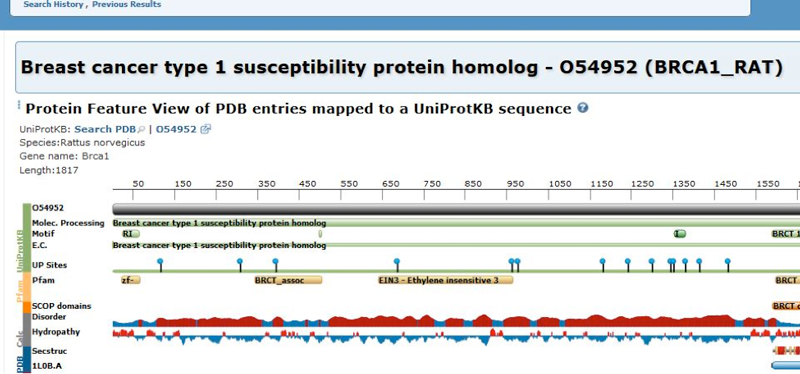
Among other things, the Protein Feature View page provides a graphical summary of a full-length protein sequence from UniProt and how it relates to PDB entries.
The PDB to UniProt mapping is based on the data provided by the Structure integration with function, taxonomy and sequence (SIFTS) initiative.
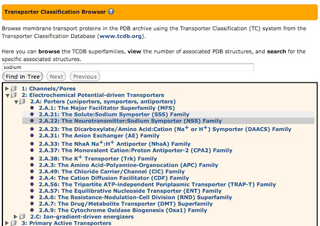
The Transporter Classification Database
(http://www.tcdb.org) organizes membrane transport
proteins using
the Transporter Classification (TC) system.
Similar to the Enzyme Commission (EC) system, it
incorporates both functional and phylogenetic information.
RCSB PDB's Browse Database option explores the PDB archive using different hierarchical trees. Each browser opens up to the top level in the hierarchy, and clicking on the arrow icons expands the respective nodes. A text box can also be used for related keyword searching. Clicking on the node's folder or name retrieves all PDB entries associated with the selected node.
Browsers are available to explore a variety of features, including GO Terms (for biological process, cell component, or molecular function), Enzyme Classification, Source Organism, Genome Location, Medical Subject Headings (MeSH), SCOP, CATH, ATC, Transporter Classification, and Protein Symmetry. Each browser can be selected from the corresponding tab.
This functionality is also an option in Advanced Search to combine these types of searches with queries for other specific types of data.
4HHB.A vs 4HHB.B using jCE click to launch