| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| April 2017 | 575,554 | 1,144,078 | 2597.41 GB |
| May 2017 | 543,609 | 1,120,077 | 2289.27 GB |
| June 2017 | 419,900 | 969,889 | 2058.14 GB |
Website statistics are calculated using AWStats
The same protein may display conformational flexibility in different experimental structures. Information regarding structural variation in similar sequences is now available on the Sequence Similarity tab of the Structure Summary Page through the integration of data from the PDBFlex database (pdbflex.org).
PDBFlex explores the intrinsic flexibility of protein structures by analyzing structural variations of the same protein across the archive. This allows for the easy identification of regions and types of structural flexibility present in a protein of interest. Structures of protein chains with identical sequences (sequence identity > 95%) are aligned, superimposed and clustered.
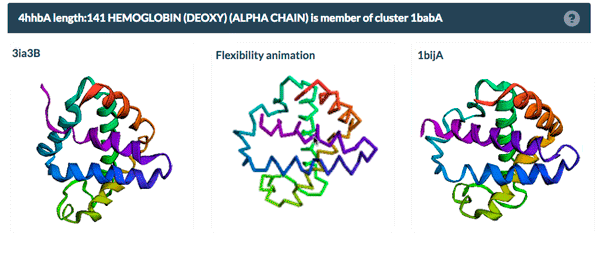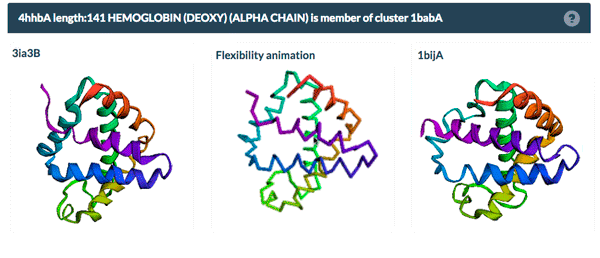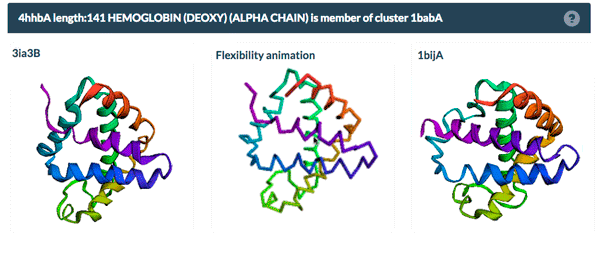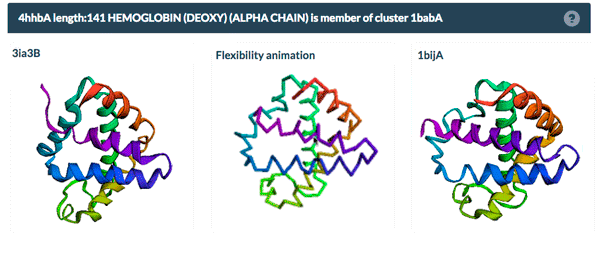
The example above shows the PDBFlex link from the Sequence Similarity tab for entry 4hhb and a view from the PDBFlex page for cluster 1babA of which the α-chain of entry 4hhb is a member. Here, the two structures within the cluster that have the highest Cα RMSD are displayed as well as a transformation between the two that helps to visualize the maximal flexibility in the cluster.
The identification of similar sequences in this report is based on the clustering used by RCSB PDB.
 Users can now access all RCSB.org data and services over the HTTPS (“secure”) protocol. This includes:
Users can now access all RCSB.org data and services over the HTTPS (“secure”) protocol. This includes:
Structure Summary Pages
3D Viewers
Search Results
REST Services
No changes need to be made to utilize this feature. While HTTPS is offered, it is not required. HTTP will be supported for the foreseeable future.
For ongoing technical updates, visit status.rcsb.org or subscribe to the RCSB PDB Status Update mailing list.
The model files in the PDB FTP archive have been updated to V5.0 of thePDBx/mmCIF dictionary.Both mmCIF and XML formats have been updated. PDB format files do not contain all of the remediated information, as PDB format is a legacy format. These files were provided previously for community review.
The changes to V5.0 include:
The complete list of changes can be found at wwPDB.org.