Education Corner
Gaming Structural Biology for General Audiences
These two outreach initiatives are making structural biology fun and accessible for broad audiences while highlighting specific experimental techniques.
XFEL Crystal Blaster: an educational game by Fiacre Kabayiza and Bill Bauer, Ph. D. |
Deep Learning, Citizen Science & Puppies: The path towards segmentation of a cryoEM dataset of a virus infected cell
by Michele Darrow, Ph.D.

Michele Darrow, Ph.D.
Michele Darrow is a Development Scientist at TTP Labtech and a Visiting Scientist at Diamond Light Source, the UK’s national synchrotron. She received her Ph.D. in structural biology from Baylor College of Medicine in Houston, TX. She is a developer of a semi-automated structural biology segmentation software named SuRVoS Workbench and co-investigator on multiple projects combining segmentation of structural biology data and citizen science. She is also developing a new semi-automated sample preparation system for cryo-electron microscopy called Chameleon.
Spare 10 minutes to make science leap forward with Science Scribbler: Virus Factory at www.diamond.ac.uk/zooniverse
The human visual system is very sophisticated in its ability to discern small differences between similar objects. Various wavelengths of light bounce off the objects in front of you, then collide with and stimulate cells in your retina, forming a pattern representative of the scene. Your visual cortex is then able to interpret the scene – breakfast, a cup of coffee with a pastry, and, of course, some phone scrolling. A picture of a puppy, another one – all curled up, how adorable! Oh – wait, is that second picture actually a bagel!?
This cute meme is the epitome of difficulty for an AI system. Think about how you might define what puppies look like. Because of small differences from one example to the next, it becomes a tedious and difficult task to ensure the definition is accurate (includes all positive examples–other puppies, while excluding all negative examples–bagels). And any definition that you use may be different from the definition that I use, which introduces some uncertainty into classifications that use each definition.
For many researchers using volumetric imaging techniques to answer biological questions, this is the exact situation they find themselves in. They’ve put in the hard work to prepare their samples, collect the data and now have 3D images of the insides of cells or tissues with high enough resolution to show what’s happening. Getting to this point in the experiment generally takes the researcher weeks to months and the data produced can be terabytes in size. The next step is to inspect the data, to visually interpret the scene and ideally, to extract information from it that will inform the experimenter’s conclusions.
This manual study entails a researcher or a team of researchers tracing the outlines of the areas of interest, throughout the 3D images. This stage of the work is called segmentation and it provides measurements, counts and locations of activity happening inside the cells. As there can be many areas of interest in each cell, this vital step can be extremely time consuming. It has been estimated that it would take a single researcher over a year to do a manual comparison study between a disease state and it’s control healthy state.
Thankfully, there are some tools available that make this task less manual and faster for the researcher! For example, ‘edge detecting’ algorithms have been developed and implemented for semi-automatically finding and marking edges of areas of interest; other tools use interactive ‘shallow machine learning’ algorithms, which can be applied to interactively segment whole groups of images at a time. However, in all cases, the amount of data collected overwhelms the researcher’s ability to process and analyse every dataset.
This is where deep learning comes in. If we return to our analogy, a deep learning algorithm could learn to distinguish between puppies as a “class” of objects and bagels as a “class” of objects. However, it would take a large amount of training data – known examples of each class with an accurate description of which class they are part of. Given the cuteness of puppies and the prevalence of this meme on the internet, there is likely enough training data to answer this specific question! Unfortunately, we don’t have much training data for the researcher to use to answer their structural biology question.
To try and help with this, we have started crowd-sourcing our training data on The Zooniverse (Zooniverse.org). This is a citizen-science platform for projects spanning astronomy, the humanities, wildlife conservation, and of course, cell biology. We have curated projects using different 3D imaging styles, with different resolution scales and in different model organisms, and we’re asking citizen scientists to help us to extract information from these images and in the process provide many, many more training datasets than we could ever produce individually.
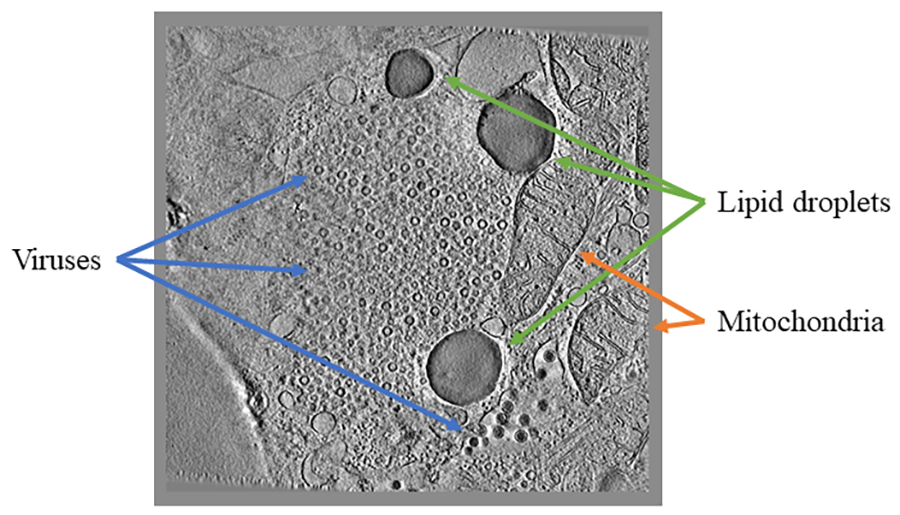
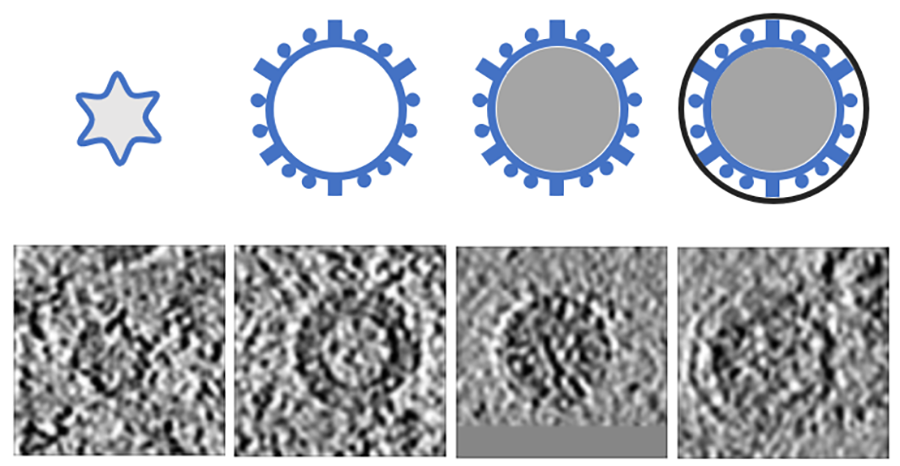
For example, in Science Scribbler: Virus Factory, we asked citizen scientists to look at sub-images from the larger dataset and mark every virus they can see (Image 1). We asked multiple citizen scientists to look at each image to ensure we would end up with a large, robust dataset that extracted as much information as possible. This resulted in tens of thousands of potential virus locations within the dataset! Each of these locations was then cut out of the dataset and re-presented to even more citizen scientists to begin removing any mis-clicks or errors, and then classifying the viruses into the various stages of the virus lifecycle (Image 2).
So far, 1,700 citizen scientists have made over 163,000 classifications to locate and identify the stages of the viruses! At the time of writing, this part of the project is 89% complete! Once, all the data are in, the next step is to do some cleaning and housekeeping with it. There are a few steps in this section; the data needs to be processed to ensure that all of the times that the citizen scientists clicked on the exact same virus are captured and joined-up in 3D. It also needs some cleaning to remove outliers and it needs a competitive algorithm to overcome cases where clear consensus was not reached. A first look at the progress made has been published.
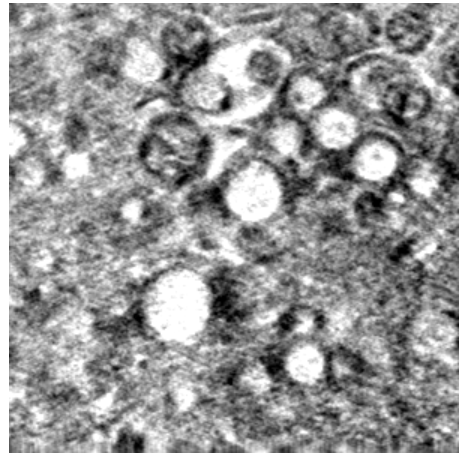
Once all of these steps are completed and shown to be working properly, the data provided by citizen scientists can be used to not only teach an algorithm how to find more viruses, but to also find other types of viruses, or even to find other similarly shaped items – like organelles in a different imaging style like soft X-ray tomography (Image 3).
This is the really interesting part of the project – how do we best join together the citizen science results with the computational algorithms to help discover the answers to biological questions? And how do we do it in such a way that it can be extended to other far-flung structural biology projects? As we move further and further into unknown territory, how do we ensure that results are accurate? It’s only by considering these questions that we can begin to harness deep learning and other computational advances to build a reasonable route to robust automated segmentation of 3D structural biology data.
If you’re interested in structural biology, segmentation and machine learning and you want to learn more or help us distinguish puppies from bagels, look for our Science Scribbler projects on the Zooniverse!
XFEL Crystal Blaster: an educational game
by Fiacre Kabayiza and Bill Bauer, Ph.D.

Fiacre Kabayiza
Fiacre Kabayiza left Africa at the age of five and relocated to Buffalo, NY. During his time as an undergraduate in the University at Buffalo’s Biomedical Science program, he participated in the BioXFEL summer internship research program. After his first summer research experience, he continued as an undergraduate researcher in Dr. Edward Snell’s laboratory at Hauptman-Woodward Medical Research Institute. It was there that Fiacre gained knowledge of protein structure determination with XFELs leading to the creation of his XFEL Crystal Blaster game and his love of computer programming. With guidance from Bill Bauer, Fiacre independently designed, programmed and distributed the XFEL Crystal Blaster game. He has since enrolled in a Master’s program in Computer Science at Georgia Institute of Technology and will graduate in 2020 with a specialization in Artificial Intelligence. In his free time, Fiacre enjoys traveling and making music.

Bill Bauer, Ph.D.
Bill Bauer is the Associate Education and Diversity Director of the BioXFEL Science and Technology Center. Dr. Bauer has a long-standing history with HWI that began with a summer student internship at HWI while attending Elmira College. After receiving a Bachelor of Science degree in Molecular Biology and a minor in Art, he entered the Interdisciplinary Graduate Program in Biological Science at the University at Buffalo. Under this program, he continued his relationship with HWI as he pursued a Ph.D. in Structural Biology focused. Dr. Bauer now manages the education and diversity programs of BioXFEL and has mentored countless undergraduates, graduate students and postdocs. He has been creating innovative and customized programming for the Scholars of the Center and the field for over 5 years.
The NSF-funded Science and Technology Center BioXFEL (www.bioxfel.org/) studies Biology with X-ray Free Electron Lasers and develops techniques for the community that facilitate the use of this new technology. BioXFEL supports several collaborative efforts that have advanced the field of XFEL science in areas of sample preparation, crystallization, target activation, sample delivery, and data collection and analysis, to name a few.
One goal of the Center is to make XFEL data collection as routine as conventional crystallography while also taking full advantage of the unique capabilities of XFEL sources. In addition to scientific contributions, BioXFEL also provides customized educational programming to educate and support a diverse group of scientists and students including educating the general public on the concepts and applications of this relatively new technology.
To help with their outreach efforts, BioXFEL intern and undergraduate researcher, Fiacre Kabayiza, and Associate Education and Diversity Director, Bill Bauer, created a game called XFEL Crystal Blaster (www.bioxfel.org/game) to educate youth on the general topics of XFEL science.

Photosystem II (PDB 4ub6)
Employing a simple, yet engaging, tap-based gameplay and a time-proven, simplistic 8-bit graphical art style, BioXFEL delivered a game that introduces young researchers to XFELs and structure determination. The game highlights three proteins that have been instrumental in initial XFEL studies and describes their structure and function - Bacteriorhodopsin (PDB structure 4x31), Photosystem II (PDB 4ub6) and Photoactive Yellow Protein (PDB 5hd5). XFEL Crystal Blaster has been used in courses at Cold Spring Harbor, demonstrated at the USA Science and Engineering Festival, featured in a museum display in the Buffalo Science Museum, and has been used in countless classrooms and outreach events including a tabletop implementation at the visitor center for the first XFEL source in Stanford University.
Players activate the XFEL by tapping on the screen as the protein crystals pass through the path of beam. Each successful crystal hit produces a diffraction pattern that adds to their data set. After enough data has been collected, the protein structure and function will be revealed. By tracking crystal hit rates, players can replay and compete for the highest score. There are currently three levels each with increasing difficulties. As the player progresses through the game, they will learn of the importance of various proteins and how researchers use these structures to learn more about the world around us.
XFEL Crystal Blaster employs a game-based learning strategy that is designed to balance the subject matter with the gameplay experience. The primary objective is to deliver information about XFEL protein structure determination in a fun and easy to understand format. Educational content is delivered at two key places. First, the scrolling introduction screen explains the basic premise of protein crystallography. Second, upon completion of each level, the player is presented with a rotating protein structure and short description of the importance of the protein and the significance of studying this particular target.
Players should come away from this game with basic knowledge of how scientists solve protein structures, the functions of these particular proteins, and why studying them is important.
XFEL Crystal Blaster is easily accessible and is available on the BioXFEL website, Apple iTunes, and Google Play. So far, the game has been downloaded hundreds of times, with thousands of proteins determined using the online version.