Education Corner
Using the Protein Data Bank in the College Classroom
by Bonnie Hall, Ph.D., Grand View University, Iowa

Bonnie Hall, Ph.D.
Bonnie Hall is an Associate Professor of Chemistry at Grand View University, Iowa, where she is always looking for new active learning strategies to engage students with biochemistry. Her first experience with the Protein Data Bank was through implementing an Abl-kinase-focused, semester-long laboratory curriculum. One component required students to download and analyze PDB files using a limited number of tools in PyMOL. Together, students explored how Chronic Myeloid Leukemia (CML) patients develop resistance to the drug Gleevec by understanding the relevant drug-protein interactions visualized in PDB structures.
Bonnie now participates in the BioMolViz project, allowing her to explore in more depth how to best teach and assess the molecular visualization found in her courses. She is a new member of the Biochemistry Authentic Scientific Inquiry Laboratory (BASIL) course-based undergraduate experience, where student learn to think like scientists as they study proteins with known PDB structures but no known function. Bonnie is also a faculty sponsor for a CREST Biomolecular Modeling team. Enthusiastic student responses to what started as a simple PDB-focused exercise in one course has expanded to molecular exploration of PDB files as a significant educational tool in many of her courses.
Bonnie can be reached at bhall @ grandview.edu.The GOB or Health Sciences Classroom
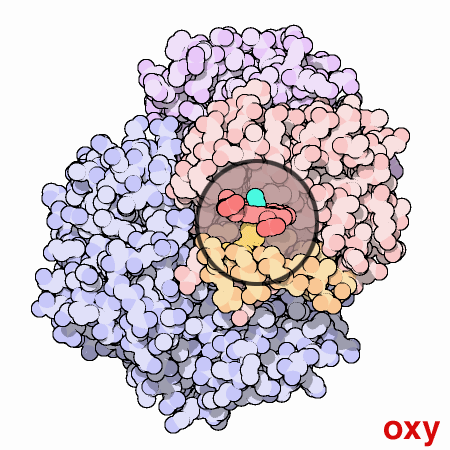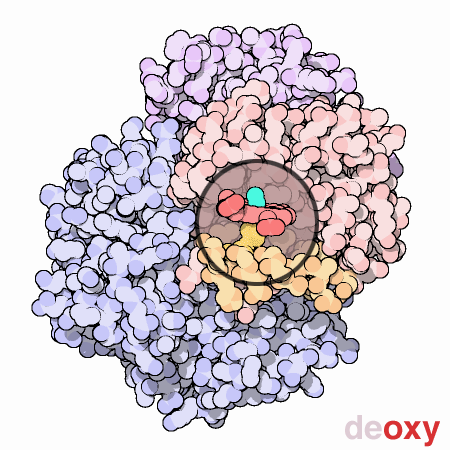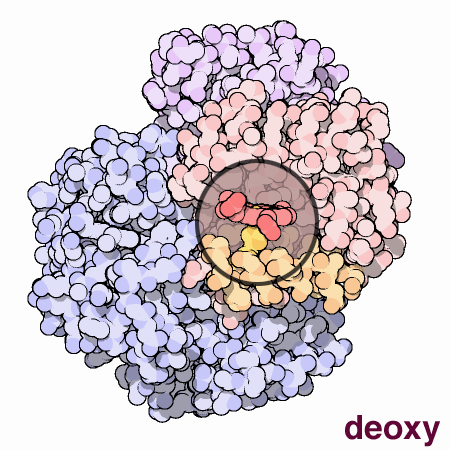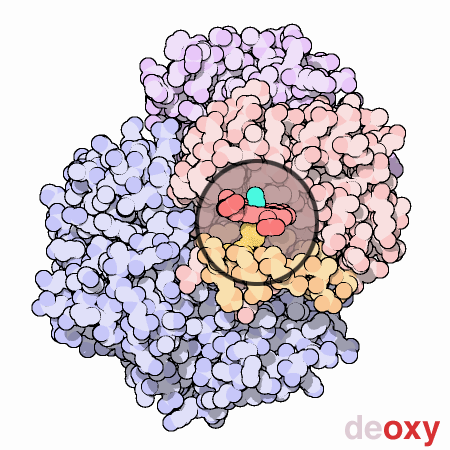
A one- or two-semester course covering General, Organic and Biochemistry (GOB) is frequently required for students entering programs in nursing and other health-related careers. Articles in the PDB-101 Molecule of the Month collection allow students in the GOB classroom to engage with protein structures in a limited but informative way. One example is the Molecule of the Month article about hemoglobin (1). Understanding the structure of hemoglobin allows students to understand how the cooperative effect in an oxygen binding curve is connected to the treatment of asthma, the molecular basis of sickle cell disease, and why carbon monoxide is such a deadly gas. Understanding drug action is an important part of modern medicine, and the How Do Drugs Work? flyer (2) provides a good introductory explanation. Other topics covered by introductory material include the transferases responsible for generating ABO blood types (3), the role of serum albumin in transport (4), and a poster on Insulin and Diabetes (5). All these materials expose students to the modern biochemistry that underlies our understanding of many physiological processes.
The Biochemistry Classroom
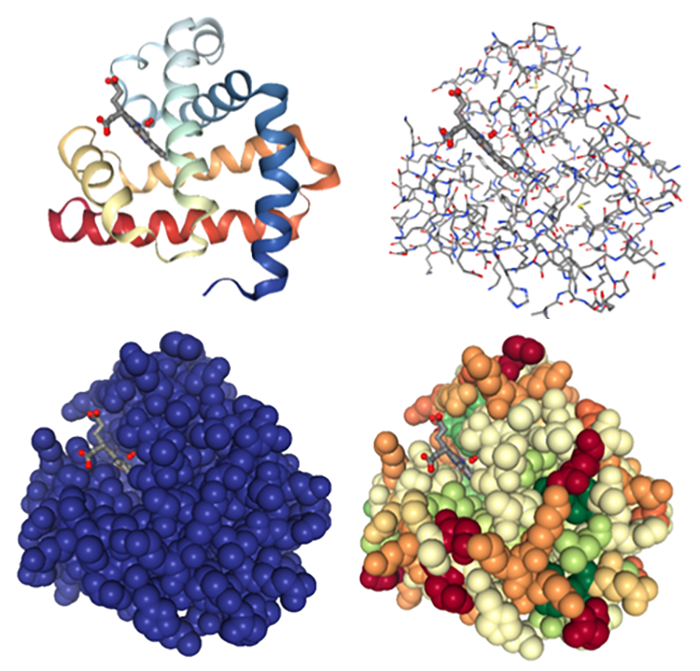
Figure 2. 3D images of myoglobin using a variety of structure views in NGL, PDB structure 1mbo (6).
Contemporary biochemistry textbooks are full of images from the PDB, with the PDB ID provided for each image. A simple activity to help students understand protein folding, structure and function is have them select one or two of the proteins from the chapter being discussed. Utilizing the various viewing tools in the “3D View” section of a given PDB entry, it is easy to explore protein secondary structure, amino acid composition, the usefulness of various protein viewing modes, and properties such as hydrophobicity. Viewing a substrate bound in an active site allows for rich discussions about role of intermolecular forces in substrate binding. If viewed using a surface view of the protein it nicely demonstrates just how specific the fit is for many binding pockets. The “Ligand Interaction” view allows critical interactions between the protein and ligand to be visualized, and helps explain how catalysis occurs.
When studying enzyme kinetics and inhibition, using a PDB file to explore enzyme function also helps students understand what appears to be a mutually exclusive set of conditions—the substrate of an enzyme also functioning as an allosteric modifier of its activity. The Structures of the Citric Acid Cycle flyer (7) provides a simple overview of the steps involved, and is useful as a pre-reading item when discussing this pathway. The Pyruvate Dehydrogenase Complex Molecule of the Month entry (8) provides a good explanation of how a large macromolecular structure is solved, and how this complex functions.
A Biomolecular Modeling Course
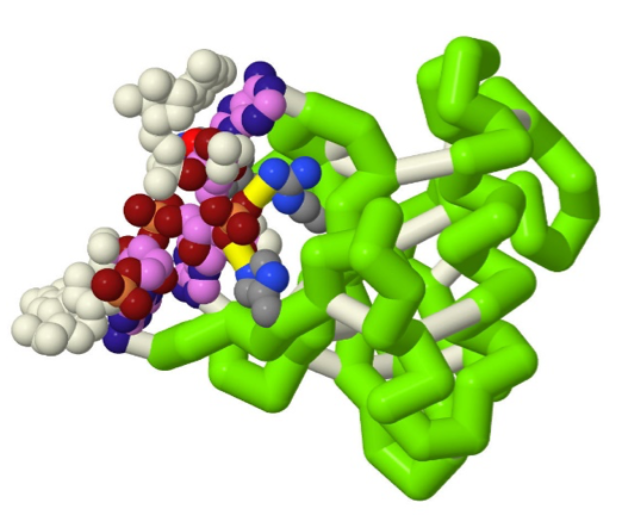
Figure 3. Jmol image showing the interaction of HigB protein and its mRNA substrate, generated by a CREST Team at Grand View University, PDB structure 4zsn (10).
When studying enzyme kinetics and inhibition, using a PDB file to explore enzyme function also helps students understand what appears to be a mutually exclusive set of conditions–the substrate of an enzyme also functioning as an allosteric modifier of its activity. The Structures of the Citric Acid Cycle flyer (7) provides a simple overview of the steps involved, and is useful as a pre-reading item when discussing this pathway. The Pyruvate Dehydrogenase Complex Molecule of the Month entry (8) provides a good explanation of how a large macromolecular structure is solved, and how this complex functions.
The Biochemistry Laboratory
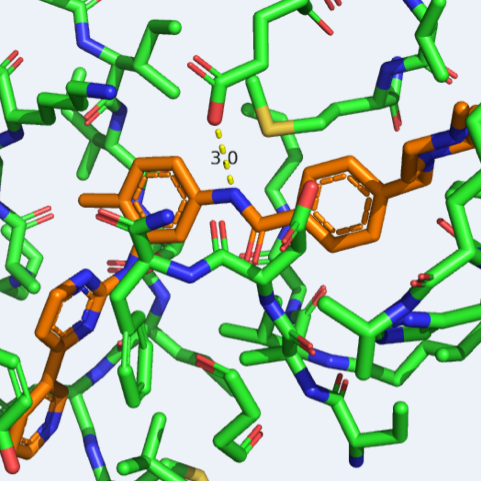
Figure 4. Looking for appropriate hydrogen-bonding sites between Gleevec and the Abl kinase, PDB ID 1iep (12).
It has become increasingly popular to offer semester-long laboratory sequences, with a series of connected experiments that allow students to learn biochemical methods in a more research-like flow. Protein overexpression and purification along with some form of enzyme kinetics are frequent components of such a series of experiments. Whether a laboratory course consists of a connected series or a set of independent experiments, utilizing the PDB can help students understand those experiments at a deeper level. By utilizing PyMOL or Chimera to explore relevant PDB structures, students identify critical molecular details of enzyme function and tie it back to foundational biochemistry concepts. An example of such an exercise is part of the laboratory sequence designed by Vogel Taylor et al. (11). Students work with the Abl kinase, performing protein over-expression, protein purification and enzyme kinetics experiments. In parallel, they use PyMOL to explore both the action of various inhibitory drugs on Abl kinase function, as well as the development of drug resistance in chronic myeloid leukemia patients. Connecting the molecular events to events at the level of the whole organism helps students understand just how the action of biomolecules underlies biological, physiological and organismal outcomes.
Upper Division Electives in Chemistry
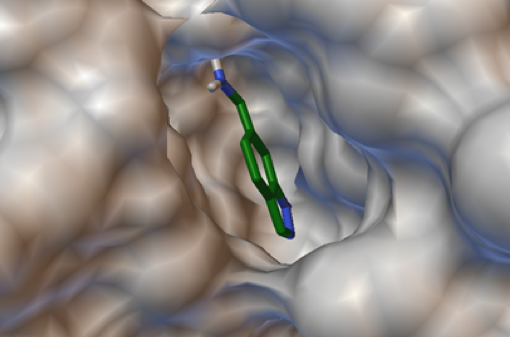
Figure 5. Student-generated image showing the docking of a re-designed drug into the active site of its target, PDB structure 2vta (15).
An upper division elective that can be popular with both biology and chemistry majors is medicinal chemistry. A project-based approach (13, 14) allows students to get hands-on experience with basic principles of drug action and drug design. Extensive use of PDB files is a natural outcome of such a format, and often include asking students to re-design an existing drug and to dock it back into a known target structural file from the PDB.
Structural bioinformatics can also be taught as an upper-division elective in chemistry. Students can utilize a variety of public databases to understand protein structure function relationships, including BLASTp, DALI, Pfam, PANNZER and RCSB PDB. Students explore how different types of information (gene sequence, protein sequence, protein tertiary structure, phylogenetic relationships) help researchers determine the function of novel proteins. The Biochemistry Authentic Scientific Inquiry Laboratories (BASIL) consortium provides one example of a public-facing curriculum that can be utilized to run or design such a course (16).
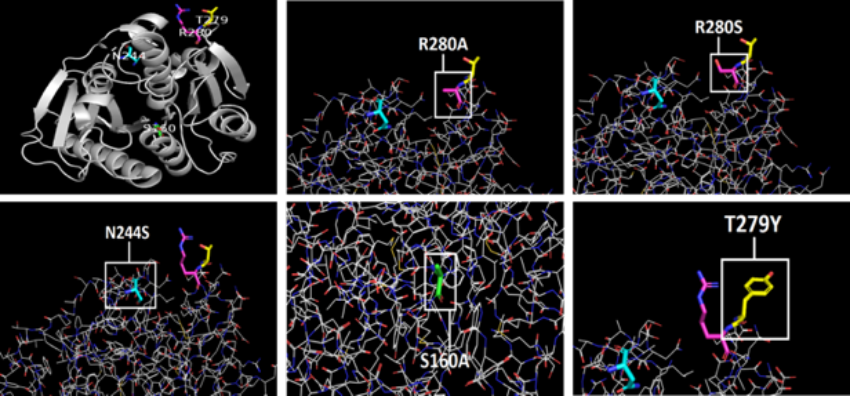
The PDB is also very useful for students utilizing proteins and/or enzymes in their research. For example, students selecting mutants to make when studying protein function (17) can use PDB structures in conjunction with visualization software such as PyMOL or Chimera. Although this will not provide information about how a protein might be affected conformationally, it does allow students to visualize the specific region in a protein that would be impacted by a mutation. Software such as Rosetta or Foldit are more complex to use, but allow students to examine potential effects on protein folding.
Figure 6. Research student images (17) demonstrating a site-directed mutagenesis experiment and why various mutants were selected, PDB structure 6eqe (18).
Personal Experiences Using the PDB
As a teaching tool, I have found RCSB PDB to be an invaluable resource. Structure-function relationships are a critical part of teaching Biochemistry at any level. Molecular visualization helps students understand those relationships, but is a challenging skill for them to master. Being able to utilize PDB data in a variety of ways has allowed me to engage students much more deeply with foundational concepts in Biochemistry. Try using the RCSB PDB with a class—I’m always pleasantly surprised at how much students enjoy PDB images, and how much it helps them understand the relevant concepts.
Citations
- Dutta, S., and D. Goodsell. “Hemoglobin.” PDB Molecule of the Month. 2003. DOI:10.2210/rcsb_pdb/mom_2003_5
- PDB-101 “How Do Drugs Work?”
- Goodsell, D. “ABO Blood Type Glycosyltransferases.” PDB Molecule of the Month. 2012. DOI:10.2210/rcsb_pdb/mom_2012_12
- Goodsell, D. “Serum Albumin.” PDB Molecule of the Month. 2003. DOI:10.2210/rcsb_pdb/mom_2003_1
- PDB-101“Insulin and Diabetes”
- Phillips, S.E. “Structure and Refinement of Oxymyoglobin at 1.6 A Resolution.” J. Mol. Biol. 1980. 142, 531-554. DOI:10.1016/0022-2836(80)90262-4
- PDB-101 “Structures of the Citric Acid Cycle”
- Goodsell, D. The “Pyruvate Dehydrogenase Complex.” PDB Molecule of the Month. 2012. DOI:10.2210/rcsb_pdb/mom_2012_9
- NSF Award Search: Award# 1725940 – DUE: “CREST3: Engaging Undergraduates in the Community of Science through Modeling.” 2017
- Schureck, M.A., A. Repack, S.J. Miles, J. Marquez, and C.M. Dunham. “Mechanism of Endonuclease Cleavage by the HigB Toxin.” Nucleic Acids Res. 2016. 44, 944-53. DOI:10.1093/nar/gkw598
- Taylor, E.V., J.A. Fortune, and C.L. Drennan. “A Research-inspired Laboratory Sequence Investigating Acquired Drug Resistance.” Biochem. Mol. Biol. Educ. 2010. 38, 247-52. DOI:10.1002/bmb.20384
- Nagar, B., W. Bornmann, P. Pellicena, T. Schindler, D.R. Veach, W.T. Miller, B. Clarkson, and J. Kuriyan. “Crystal Structures of the Kinase Domain of c-Abl in Complex with the Small Molecule Inhibitors PD173955 and Imatinib (STI-571).” Cancer Res. 2002. 62, 4236-4243
- Hall, B.L., K. D. Watson, and T. Covey. “Resources for Teaching Project-Based Undergraduate Medicinal Chemistry Courses.” Technology Integration in Chemistry Education and Research (TICER) 2019. Chapter 9, 131-142. DOI:10.1021/bk-2019-1318.ch009
- Tantillo, D.J., J.B. Siegel. C.M. Saunders, T.A. Palazzo, P.P. Painter, T.E. O’Brien, N.N. Nuñez, D.H. Nouri, M.W. Lodewyk, B.M. Hudson, S.R. Hare, and R.L. Davis. “Computer-Aided Drug Design for Undergraduates.” .J Chem. Educ. 2019 96, 920-925. DOI:10.1021/acs.jchemed.8b00712.
- Wyatt, P.G., A.J. Woodhead, V. Berdini, J.A. Boulstridge, M.G. Carr, D.M. Cross, D.J. Davis, L.A. Devine, T.R. Early, R.E. Feltell, E.J. Lewis, R.L. McMenamin, E.F. Navarro, M.A. O'Brien, M. O'Reilly, M. Reule, G. Saxty, L.C. Seavers, D.M. Smith, M.S. Squires, G. Trewartha, M.T. Walker, and A.J. Woolford. “Identification of N-(4-piperidinyl)-4-(2,6-dichlorobenzoylamino)-1H-pyrazole-3-carboxamide (AT7519), a Novel Cyclin Dependent Kinase Inhibitor Using Fragment-based X-ray Crystallography and Structure Based Drug Design.” J. Med. Chem. 2008. 51, 4986-99. DOI:10.1021/jm800382h.
- McDonald A.R., H.J. Bernstein, S.C. Daubner, A. Goodman, S.M. Irby, J.R. Koeppe, J.L. Mills, S.F. O’Handley, M.J. Pikaart, R.A. Roberts, A. Sikora, and P.A. Craig. “BASIL Biochemistry Curriculum.” 2019. https://basilbiochem.github.io/basil/.
- Duplan, A., and B. L. Hall “More Efficient Breakdown of PET Plastic by Modifying the PETase Enzyme.” The FASEB Journal 2019. 33:1_supplement, 472.3-472.3.
- Austin, H.P., M.D. Allen, B. S. Donohoe, N.A. Rorrer, F.L. Kearns, R.L. Silveira, B.C. Pollard, G. Dominick, R. Duman, K. El Omari, V. Mykhaylyk, A. Wagner, W.E. Michener, A. Amore, M.S. Skaf, M.F. Crowley, A.W. Thorne, C.W. Johnson, H.L. Woodcock, J.E. McGeehan, and G.T. Beckham. “Characterization and Engineering of a Plastic-degrading Aromatic Polyesterase.” Proc. Natl. Acad. Sci. USA. 2018. 115, E4350-E4357. DOI:10.1073/pnas.1718804115.