Data Exploration Services
2020 RCSB.org Statistics
RCSB PDB's website at rcsb.org was visited each month by an average of ~717,000 unique visitors hosting millions of visits in 2020. Traffic is tracked using AWstats.
| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2020 | 666,572 | 2,520,136 | 8,300.07 GB |
| February 2020 | 678,356 | 2,546,039 | 7,371.69 GB |
| March 2020 | 848,758 | 2,979,171 | 7,337.97 GB |
| April 2020 | 813,119 | 2,930,280 | 9,489.47 GB |
| May 2020 | 780,867 | 2,782,238 | 8,723.20 GB |
| June 2020 | 693,141 | 2,491,024 | 8,874.05 GB |
| July 2020 | 707,006 | 2,553,617 | 10,566.80 GB |
| August 2020 | 682,088 | 2,162,671 | 4,080.59 GB |
| September 2020 | 686,187 | 2,285,846 | 2,679.69 GB |
| October 2020 | 666,900 | 2,106,459 | 3,355.89 GB |
| November 2020 | 695,906 | 2,365,871 | 4,189.16 GB |
| December 2020 | 667,787 | 2,323,819 | 3,514.68 GB |
Documentation for New and Improved APIs
Recently-introduced Search and Data APIs offer comprehensive functionality and high performance.
The new Search API allows users to run queries across RCSB PDB Search Services and retrieve a list of relevant identifiers such as PDB IDs, entity IDs, assembly IDs, and more.
Documentation is available to help users learn the concepts and syntax behind the new services. Use this documentation to display all of the details of a specific API request, and then run the search request and to see the response.
wwPDB News: Time-stamped Copies of the PDB Archive
A snapshot of the PDB archive (ftp://ftp.wwpdb.org) as of January 5, 2019 has been added to ftp://snapshots.wwpdb.org and ftp://snapshots.pdbj.org. Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20210105 includes the structure and experimental data for the 173,005 PDB entries available at that time. Atomic coordinate and related metadata are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot is 822 GB.
Scientific Software Developers and Postdocs: Join Our Team

RCSB PDB team pictured at the January 2020 Cloud Technologies Best Practices hosted by the Institute for Quantitative Biomedicine at Rutgers, The State University of New Jersey.
For more, visit our Careers page or Contact Us with questions.
Sequence Alignments in Search Results
Use the Advanced Search>Sequence option to search protein, DNA, or RNA sequences using mmseqs2 software. A description of this functionality is available.
The search results include a graphic display of sequence alignments. Please adjust the "Display Results as" menu to "Polymer Entities". Zoom using the mouse scroll (wheel) or the mousepad/touchpad to show the amino acid sequence and mismatches. 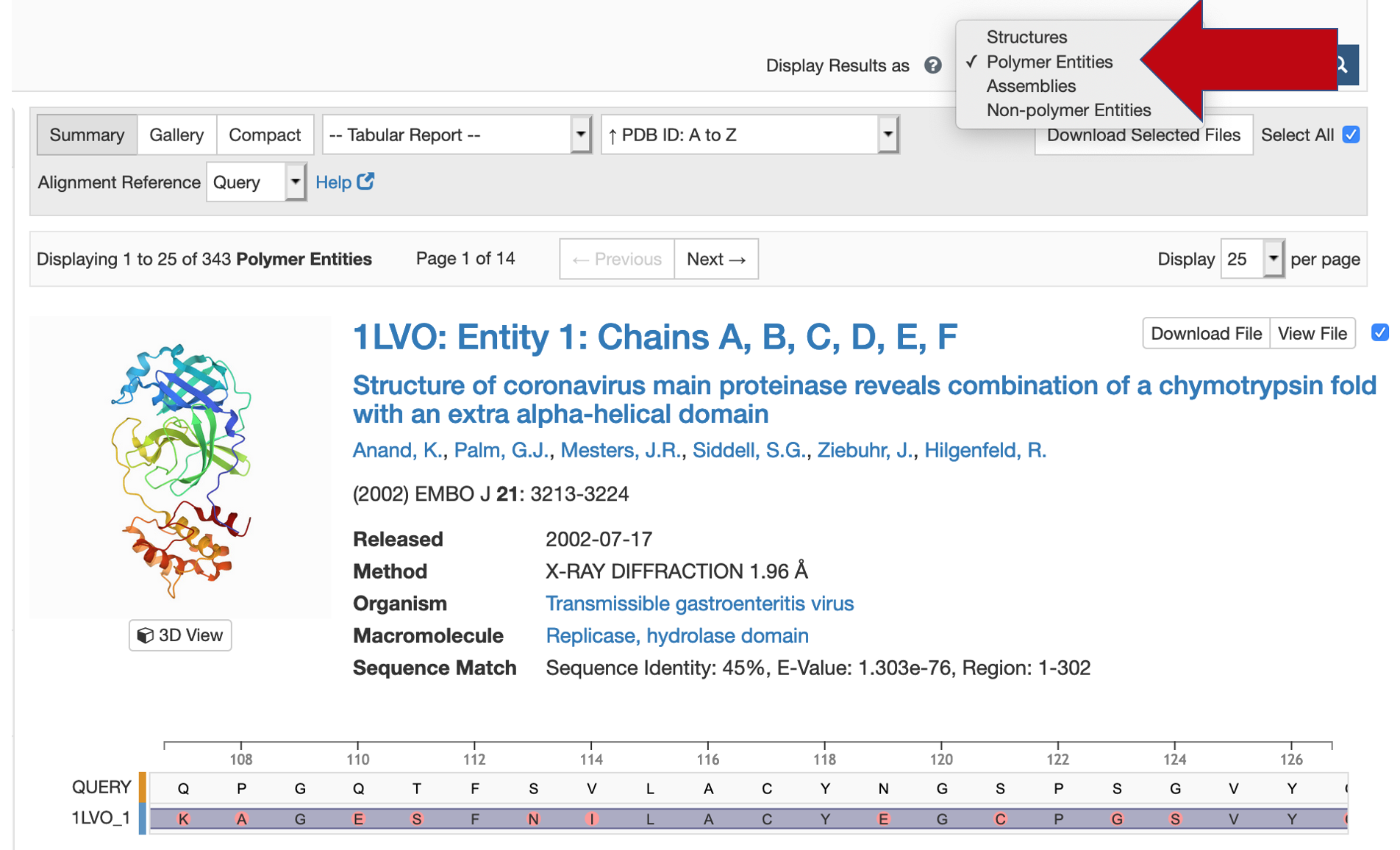
Zoom using the mouse scroll (wheel) or the mousepad/touchpad to show the amino acid sequence and mismatches. Help documentation is available.
Advanced Search Query Builder can be used combine these Sequence Searches with other types of searches using Boolean operators (AND/OR/NOT):
- Attribute searching: specific fields or full-text search across all fields, such as ID, deposition information, entry features, and experimental information
- Sequence searching: based on a FASTA sequence (with E-value or % Identity cutoffs);
- Sequence Motif searching: find short sequence patterns in PDB structure FASTA sequences using Simple, PROSITE, or RegEx syntax
- Structure Similarity searching: based on an existing Chain or Assembly of a PDB structure
- Chemical Search: find chemical components by Formula or descriptor
Building Advanced Searches
Use Advanced Search>Attribute Search to query specific fields in the database or to perform a full-text query across all fields.
The database maintains a long list of attribute fields. Users can browse the pull-down menu of options, or search to find relevant categories. For example, type resolution to find the corresponding database attributes that can be queried.
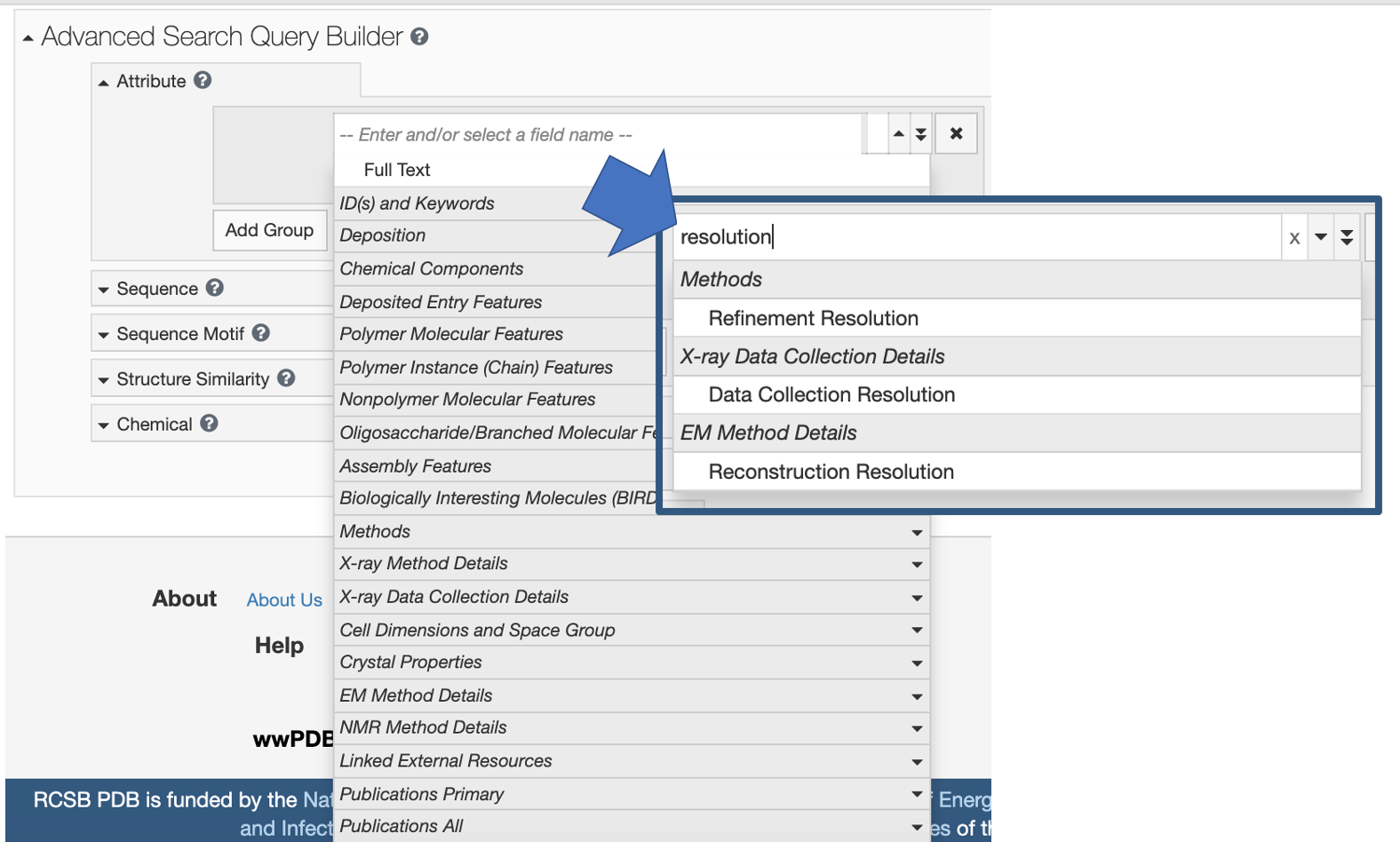
Entering the word resolution highlights the related attributes that can be searched.
Explore Improved Carbohydrate Data
A new data representation for carbohydrates in PDB entries and reference data improves the Findability and Interoperability of these molecules in macromolecular structures.
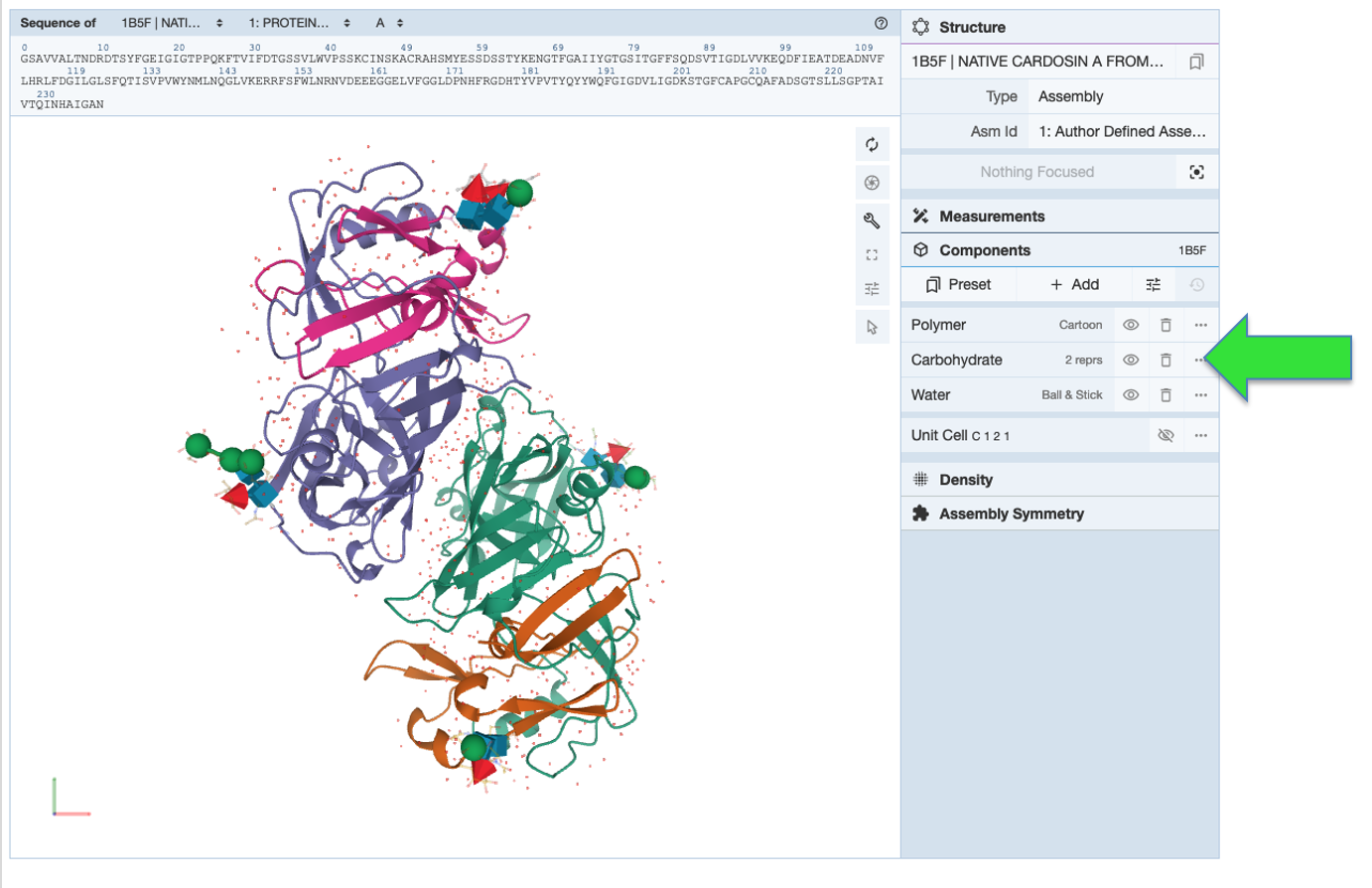
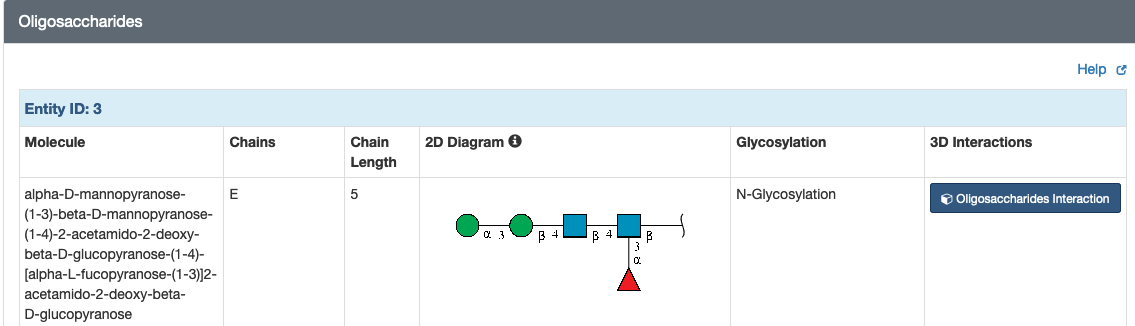
Access oligosaccharide Information from Structure Summary pages (example: 1b5f), including names, 2D Symbol Nomenclature For Glycans (SNFG) images, glycosylation sites, and 3D interaction view. BIRD molecules for entries containing commonly-known di-, tri- saccharides or blood antigens that have BIRD definitions are also listed.
Structure Summary page focused on oligosaccharide Information
The Mol* 3D view options displays 3D SNFG representations for carbohydrates for easy identification of sugars and their neighboring environment.
Mol* 3D view with oligosaccharides shown using same color scheme as 2D SNFG diagram
From the top bar Basic Search, find oligosaccharide containing entries by a specific glycosylation site or by an exact linear descriptor provided in the PDB files.
Use Advanced Search to find entries with glycosylation in the Polymer Molecular Features or entries with Oligosaccharide Features by searching any of these attributes: linear descriptors, systematic name, monosaccharide composition, etc.
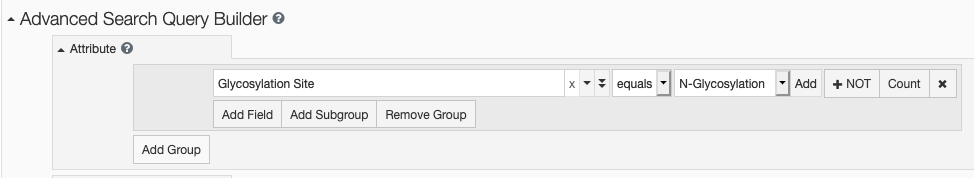
Advanced Search options
Oligosaccharide features from the return of search results can be created and downloaded using pre-defined Oligosaccharide or Custom tabular report.
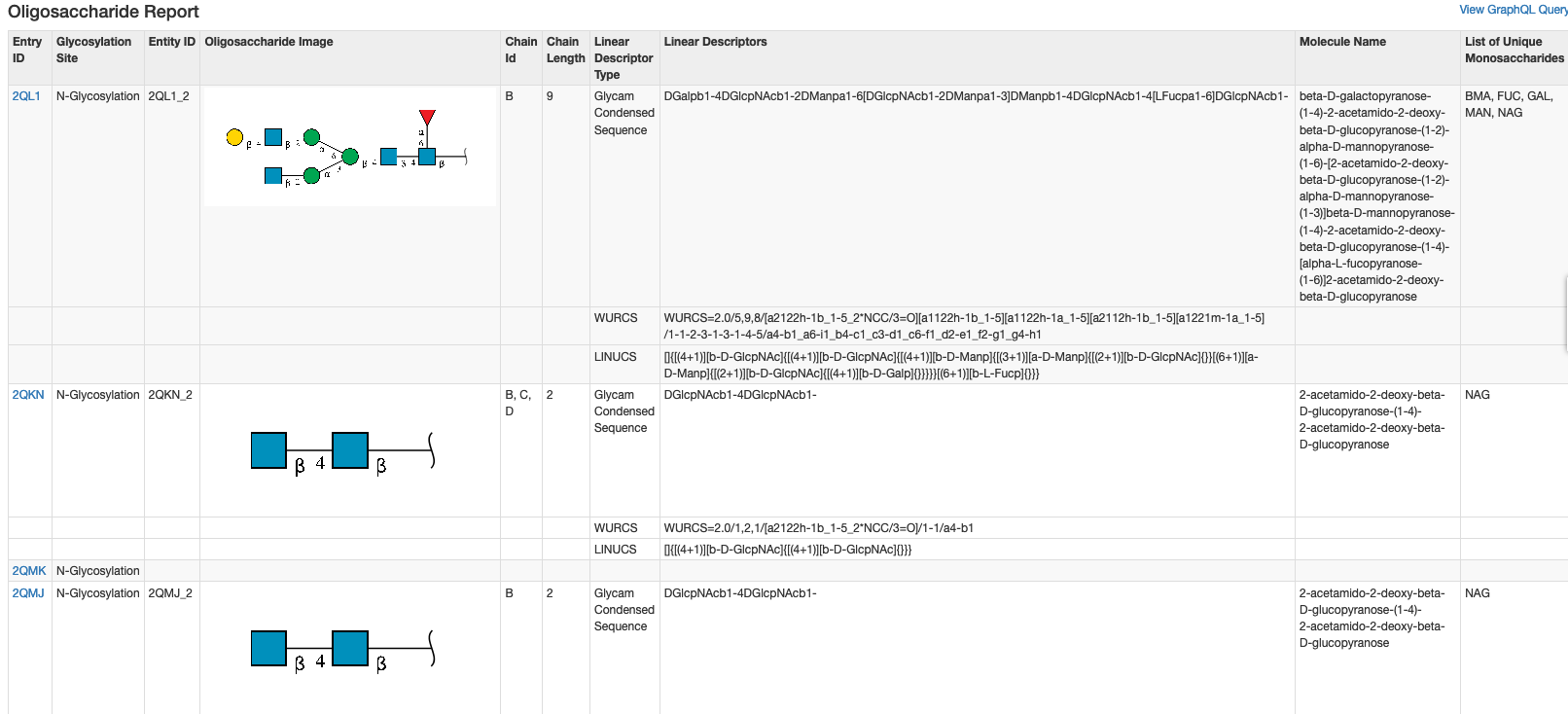
Items in the Oligosaccharide Report can also be used to generate a Custom Report