Education Corner
Exploring the Structure-Function Relationship with Digital and Physical 3D Models of Proteins
by Maria Voigt, RCSB PDB

Maria Voigt is the Senior Outreach Coordinator at the RCSB Protein Data Bank, based at Rutgers, The State University of New Jersey. She focuses on visual aspects of educational outreach, using illustration, animation, and graphic design that are made freely available at PDB-101. Her work has appeared in The New York Times and recognized by the Wellcome Trust, FASEB, the American Public Health Association, and others. Her video What is a Protein has been viewed more than 1.6 million times since release in 2017 and translated into multiple languages. Maria also coordinates hands-on science outreach events that get students excited about the structure and function of proteins.
The data in the Protein Data Bank are the result of experimental exploration of 3D structures of proteins. When the structure is revealed, the scientists gain insights into how the arrangement of the elements and interactions between them allow the protein to fulfill its function. This knowledge advances our understanding of biology and can be used in drug design and bioengineering. This knowledge can also be used in educational settings for the students to gain insight into the patterns of structural features and interactions that power life.
The most common visual medium through which students get acquainted with the protein structures and their functions are 2D depictions in textbooks. These depictions have a few shortcomings. First of all, they often offer arbitrary shapes to stand in for specific proteins. Knowing the actual 3D shape of the protein helps the students to recognize it later and build on knowledge previously acquired. Second of all, these illustrations offer only superficial view of the intermolecular interactions by using simple visuals such as matching rudimentary shapes. The covalent and non-covalent interactions guiding the function are not communicated through the 2D visual aid, even though the text accompanying the image might mention such interactions. This can create a dissonance in the information interpretation and lead to confusion. Also, in education it’s important to make connections between the disciplines. Exploring a protein in 3D along with the interactions that hold it together creates the biology-chemistry connection and gives the students a more complete view of the science.
Exploring the structure-function relationship is an integral part of the Next Generation Science Standards (NGSS). RCSB PDB websites at RCSB.org and PDB-101.rcsb.org offer tools and resources that can be used by students and teachers to explore the 3D shapes of proteins and how they enable their function. The resources vary in complexity and can be used for biology and chemistry education on middle school, high school, and undergraduate levels.
Using 3D Molecular Viewer in Guided Settings
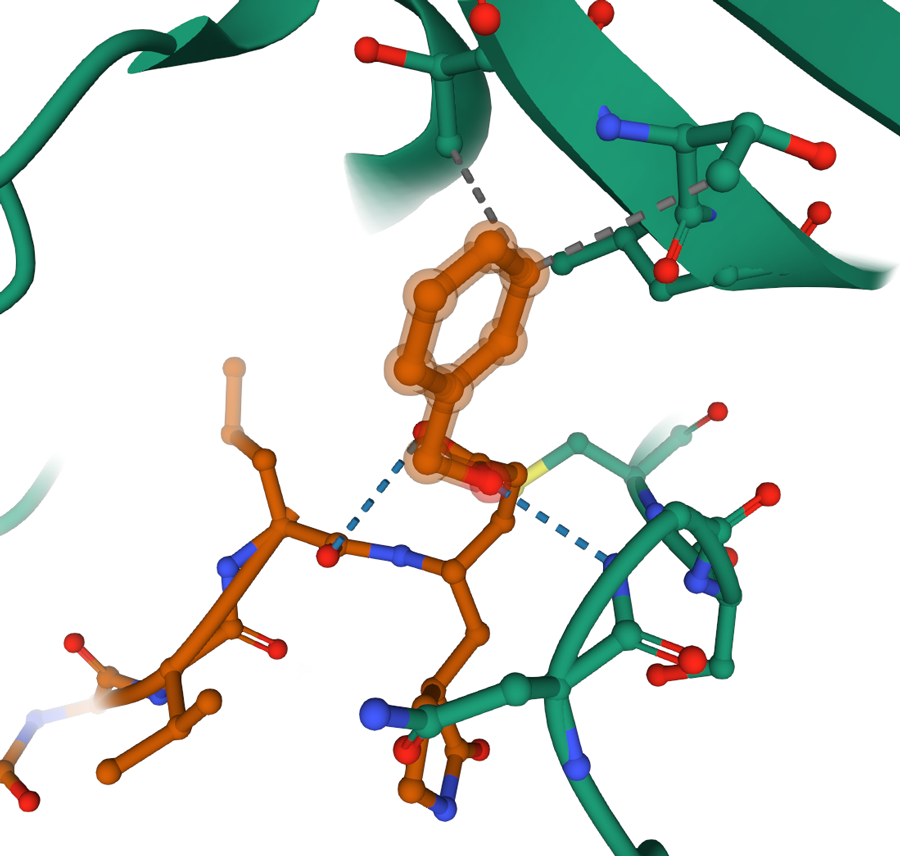
Upon clicking on a residue or a small molecule of interest, Mol* enters focus mode that allows for exploring non-covalent interactions.
An in-depth molecular exploration can be achieved using a 3D molecular viewer with advanced visualization and analysis features. Each structure in the PDB archive can be explored on rcsb.org website in 3D using the Mol* (MolStar) molecular viewer. The viewer is accessible from the “3D View” tab on each Structure Summary page. Upon opening, the viewer shows the polymer in ribbon representation and the small molecules that interact with the protein in ball and stick representation. Clicking on the molecular components focuses them in the viewport with all side chains and non-covalent interactions such as hydrogen bonds, ion bonds, hydrophobic interactions displayed. The viewer also allows extremally high granularity in selecting elements and applying a very high level of customizations for the molecular representations. The video Exploring PDB Structures in 3D with Mol* Introductory Guide highlights key Mol* features useful for exploring and analyzing 3D structures in the classroom.
The lesson plan Carrying Cargo: Exploring non-covalent interactions between proteins and small molecules in blood transport offers guided 3D exploration of the non-covalent interaction in protein transport.
The lesson plan Carrying Cargo: Exploring non-covalent interactions between proteins and small molecules in blood transport utilizes RCSB PDB features to examine the mechanisms used by serum albumin to carry fatty acids in the blood. Using Molecule of the Month, students learn about the function of serum albumin; using the rcsb.org website, students explore a fatty acid that is transported in the blood. Students then predict what structural features would a transport protein need to have in order to accommodate this molecule in the bloodstream? Students then explore the PDB structure 1e7i in 3D with Mol* to evaluate their predictions. Visualizing the chemical interactions and applying multiple molecular representations helps students to understand not only how serum albumin is structurally optimal for the job of carrying fatty acids, but also how non-covalent interactions between the protein, ligands, and surrounding water enable the transport of fatty particles in water-based blood.
Using Physical Models
In learning about structure and function, the progression of concepts is important. For example, middle school students may recognize key molecules (e.g., DNA, viruses) but not yet comprehend the chemical interactions guiding their functions. In this setting, modeling the function using a physical 3D model may be helpful. PDB-101 offers paper models and models for 3D printing that can be utilized in demonstrating the concept of structure function relationship.
Paper Models

In this Zika paper model the protein structure image is projected onto a flat icosahedron template. When the model is finalized, students can observe the repeating patterns of protein chains.
An excellent example of the structure-function relationship that can be understood without in-depth knowledge of intramolecular interactions is the viral protein capsid. The protein forms a spherical enclosure that transports viral genetic material. PDB-101 offers a variety of virus paper models. In each, the viral protein is projected onto a flat icosahedron that students cut out, fold, and glue together. Nucleic acid can be modeled using thread or pipe cleaners and inserted before the final triangle is glued into place. Through the tactile interaction with the model students not only fashion the function of the virus, but also familiarize themselves with the concept of the patterns of repeating chains in viral capsids.
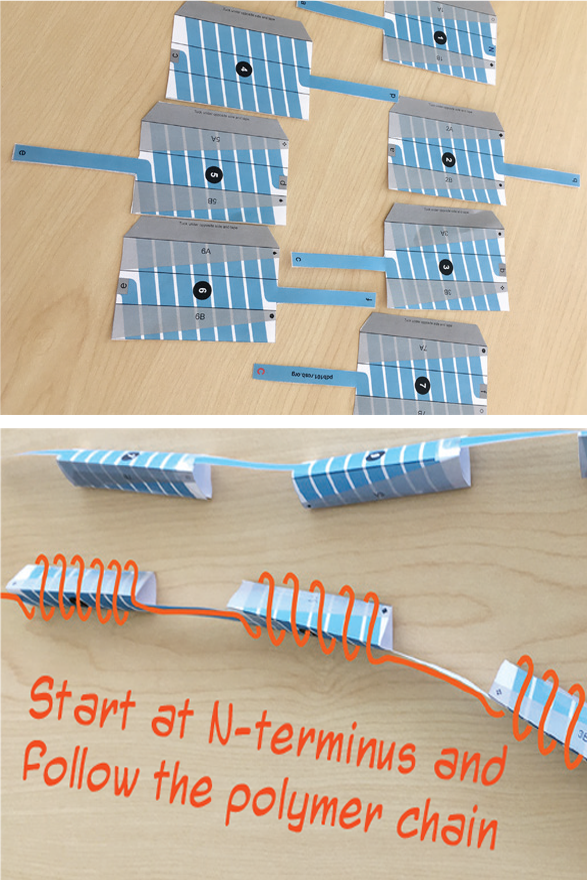
A special subset of PDB-101 paper models focuses on tactile exploration of protein structure and its levels of organization. When building these models, students start with a long strip that represents the polymer, then fold it to resemble the 3D structure.
The elements of the secondary structure are sometimes printed on the template (e.g. opioid receptor model, GFP model), or students have to ‘create’ them as they fold the model as seen in the insulin model. Students create the alpha helices by matching the CO-HN labels on the paper strip that represents the polymer to create hydrogen bonds. This helps them to get an in-depth comprehension of the forces that hold the 3D structure together.
 |
 |
 |
These models show different approaches to illustrate the secondary structure: | ||
Opioid receptor: helices are projected on parts of the template. When finalized, the single polymer line can be followed from the beginning to the end, with alpha helices indicated as 3D shapes. |
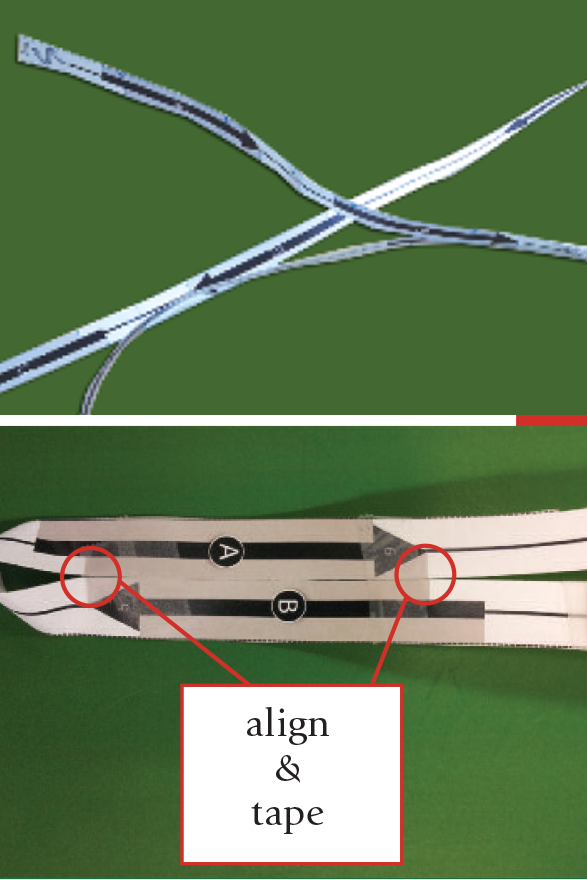
GFP: the arrows printed on the ‘polymer’ represent beta strands. When assembling the 3D model, they align to represent anti-parallel beta sheets. |
Insulin: residues and hydrogen bonds are indicated on the ‘polymer’ strip. Students roll the strip, matching the H-bond labels, to create alpha helices. |
3D Printed Models
As students get to know more molecules and systems and learn that the function of the protein is derived from its interaction with other molecules, 3D printed models can be used as visual aids. In such demonstrations it’s useful when the protein and small molecule are separate objects that can be taken apart and put together. This can be achieved by saving the protein as one file and the small molecule as another and printing them separately.
Many advanced molecular viewers offer the option of saving an .stl file that is compatible with most 3D printers. It’s important to remember that in order for the printed components to fit together, the insertable part should be scaled down to create a small offset. This can be challenging as the uniform scaling doesn't always deliver the desired results. Scaling the model in an non-uniform way can be cumbersome and require advanced modeling skills.
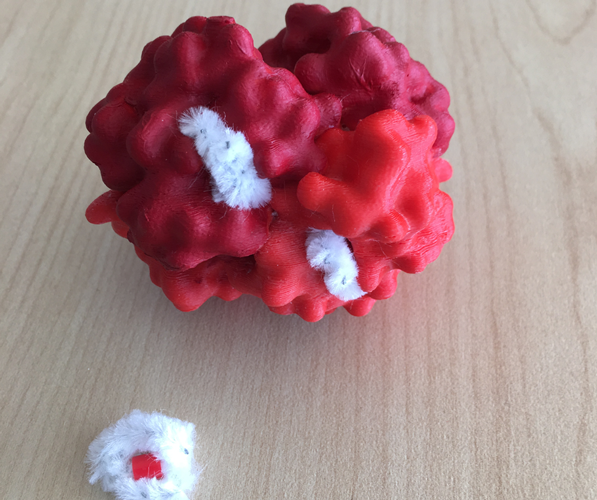
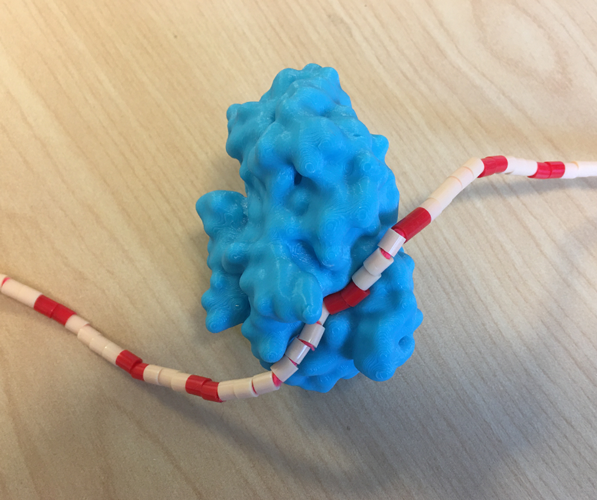
A solution is to model the small molecules from other flexible objects. The 3D print section on PDB-101 features multiple 3D printed models “interacting” with small molecule models created using pipe cleaners and 5mm melty beads.
|
|

|
| PDB-101 3D printed models of hemoglobin, alpha amylase, and serum albumin demonstrate how small molecules can be modelled from pipe cleaners and beads to easily ‘interact’ with the protein in order to demonstrate the protein-small molecule interaction. | ||

Dual extrusion printers print the model and the support system using different materials. The support system dissolves in water bath within hours or days, depending on complexity.
3D printing of proteins from the PDB is greatly enhanced by a using a dual extrusion printer. Due to irregularity of macromolecular shapes, single extrusion printers may create complex support systems that are challenging to remove. Dual extrusion printers use water-soluble material to print the support system which gets removed from the model upon immersing in a water bath.
By studying the structure-function relationship, students understand the innerworkings of objects that in turn helps to understand concepts in science and engineering. PDB-101 resources paired with RCSB.org tools offer an exciting foray into the intricacies of 3D structure.