Data Deposition/Biocuration Services and Archive Management
Deposition Statistics
In the second quarter of 2021, 3593 experimentally-determined structures were deposited to the PDB archive for a total of 7321 entries deposited in the year. Data are processed by wwPDB partners RCSB PDB, PDBe, and PDBj.
Of the structures deposited in 2021 so far, 86.4% were deposited with a release status of hold until publication; 7.8% were released as soon as annotation of the entry was complete; and 5.8% were held until a particular date. 70.8% of these entries were determined by X-ray crystallographic methods; 2.7% were determined by NMR methods; and 26.3% by 3DEM.
During the same quarter, 3334 structures were released in the PDB, including 345 SARS-CoV-2 structures. 1470 EMDB maps were released in the archive.
Future Planning: Entries with extended PDB and CCD ID codes will be distributed in PDBx/mmCIF format only
wwPDB, in collaboration with the PDBx/mmCIF Working Group, has set plans to extend the length of ID codes for PDB and Chemical Component Dictionary (CCD) ID entries in the future. Entries containing these extended IDs will not be supported by the legacy PDB file format.
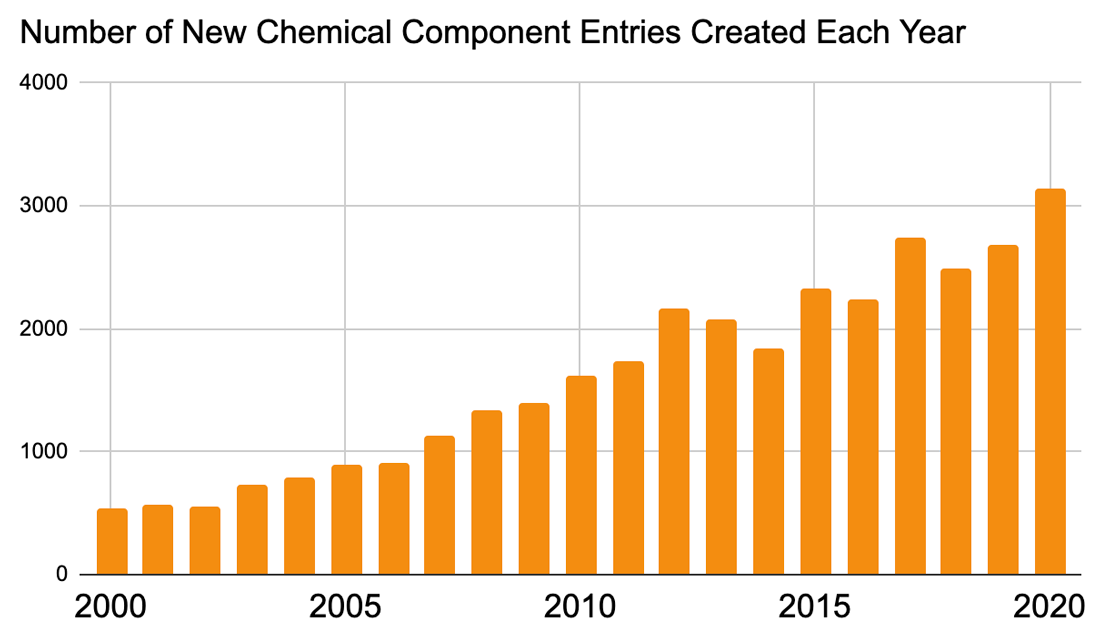
CCD entries are currently identified by unique three-character alphanumeric codes. At current growth rates, we anticipate running out of available new codes in the next three to four years. At this point, the wwPDB will issue four-character alphanumeric codes for CCD IDs in the OneDep system. Due to constraints of the legacy PDB file format, entries containing these new, four character ID codes will only be distributed in PDBx/mmCIF format. The wwPDB will begin implementation of extended CCD ID codes in 2022.
Example of map-model overlay image: EMD-30388/7CWU, SARS-CoV-2 spike proteins trimer in complex with P17 and FC05 Fabs cocktail.
In addition, wwPDB also plans to extend PDB ID length to eight characters prefixed by ‘PDB’, e.g., pdb_00001abc. Each PDB ID has a corresponding Digital Object Identifier (DOI), often required for manuscript submission to journals and described in publications by the structure authors. Both extended PDB IDs and corresponding PDB DOIs, along with existing four character PDB IDs, will be included in the PDBx/mmCIF formatted files for all new entries by Fall 2021.
For example, PDB entry 1ABC will also have the extended PDB ID (pdb_00001abc) and the corresponding PDB DOI (10.2210/pdb1abc/pdb) listed in the _database_2 PDBx/mmCIF category.
loop_
_database_2.database_id
_database_2.database_code
_database_2.pdbx_database_accession
_database_2.pdbx_DOI
PDB 1abc pdb_00001abc 10.2210/pdb1abc/pdb
WWPDB D_1xxxxxxxxx ? ?
Once four-character PDB IDs are all consumed, newly-deposited PDB entries will only be issued extended PDB ID codes, and entries will only be distributed in PDBx/mmCIF format.
wwPDB asks PDB users and related software developers to review code and begin to remove such limitations for the future.
Consistent Format for Validation and Coordinate Data
wwPDB validation reports are now provided in PDBx/mmCIF format for all new depositions in OneDep. This change makes validation data more interoperable with the PDB archival format. Data are more logically and better organized in the PDBx/mmCIF reports, and therefore more “database-friendly” than the report in XML format. PDBx/mmCIF-format validation reports for newly released and modified entries will be distributed through the PDB and EMDB Core Archives.
The new PDBx/mmCIF reports are easier to interpret. They contain a high-level summary and offer easier access to residue-level information. Data are provided at multiple levels: entity, chain-specific, and even at the individual residues. For example, it is more straightforward to obtain the total number of clashes. The corresponding validation dictionary is available at mmcif.wwpdb.org/dictionaries/mmcif_pdbx_vrpt.dic/Index. Examples of PDBx/mmCIF validation reports for X-ray, 3DEM, and NMR are publicly available at GitHub.
PDBx/mmCIF validation reports will be provided for the full PDB and EMDB archives once archival validation recalculation is performed.
wwPDB strongly recommends all PDB users and software developers adopt this format for future applications.
New pdb-l Bulletin Board
pdb-l@lists.wwpdb.org is an open discussion forum for questions and discussions with the PDB user community about protein structure, analysis, and related topics. Messages sent to pdb-l@lists.wwpdb.org will be sent to all subscribers. This bulletin board replaces the previous forum at pdb-l@sdsc.edu. Messages, including messages migrated from the previous bulletin board are archived.
Questions about PDB structures should be sent to deposit-help@mail.wwpdb.org.