Data Query, Reporting and Access
Website Statistics
Access statistics for the first quarter of 2012 are shown.
Month |
Unique Visitor |
Visits |
Bandwidth |
January |
210986 |
522651 |
982.26 GB |
February |
213862 |
526896 |
1114.34 GB |
March |
239640 |
598545 |
1004.99 GB |
New Structures Widget
 Available from the home page, the New Structures widget links to the latest structure released, the structure papers related to this release, and to the Unreleased Entry Search.
Available from the home page, the New Structures widget links to the latest structure released, the structure papers related to this release, and to the Unreleased Entry Search.
- Latest Release: The weekly update of new PDB entries can be explored and refined using the query results viewer
- New Structure Papers: A clickable list of citations associated with the entries in the latest PDB release
- Unreleased Entries: Find structures that are being processed or are on hold waiting for release. Data can be searched by PDB ID, author, title, status, sequence availability (some sequences are released in advance of the coordinates), and structural genomics project or center name.
The appearance of widgets on the RCSB PDB home page can be customized. For more, see the Winter 2012 Newsletter.
Ligand Summary Pages
 Ligand Summary Pages provide information for all of the entries found in the wwPDB's Chemical Component Dictionary.
Similar to Structure Summary pages for PDB entries, Ligand Summary Pages are organized into widgets that highlight different types of information.
Ligand Summary Pages provide information for all of the entries found in the wwPDB's Chemical Component Dictionary.
Similar to Structure Summary pages for PDB entries, Ligand Summary Pages are organized into widgets that highlight different types of information.
For example, view the Ligand Summary Page for caffeine (Ligand ID CFF) to access:
- Chemical Component Summary: an overview of the structure,including name, identifiers, synonyms, and SMILES and InCHI information
- Related PDB Entries: searches for other structures where the ligand appears as a free ligand
- Related Ligands: links to Summary Pages for similar ligands and stereoisomers, and automatically enters the ligand in the Chemical Structure Search tool to build related ligand searches
- Ligand Image: toggles between a static image and a 3D Jmol view
- Links: connects to information about the chemical component at external resources (BindingDB, HIC-Up, PDBeChem, and more) and RCSB PDB's Ligand Expo
Browse the PDB by GO Terms, EC Number, Source Organism, and More

The Browse
Database IconRCSB PDB's Browse Database feature explores the PDB archive using different hierarchical trees. Browsers are available to search for related terms and structures based upon the following classifications:
- Trees for Biological Process, Cellular Component, and Molecular Function are organized using the Gene Ontology (GO) Consortium's descriptions for gene products. PDB IDs and corresponding chain IDs have been mapped to GO terms by the SIFTS initiative.
- Enzyme Commission numbers. Search for enzymes by name, or by partial or full EC number.
- Membrane Transport proteins organized using Transporter Classification (TC) Database system.
- Source Organism, using organisms found in the NCBI Taxonomy database. These organisms are the source of the individual naturally- occuring polypeptides. Source organism assignment in the PDB is based on author/UniProtKB-specified polypeptide mapping.
- Genome Location of various organisms. The genomes represented are a subset of those in NCBI's genome database with curate sequences for genetic loci archived at Entrez Gene. The top level in the hierarchy is the organism's genome. Each genome expands into chromosomes, which in turn expand into a list of loci on the chromosomes. Each locus is a link to retrieve associated structures.
- MeSH (Medical Subject Headings) that appear in the entry's related PubMed abstract.
- SCOP description of evolutionary and functional relationships from SCOP: Structural Classification of Proteins.
- CATH clustering of proteins at four major levels from CATH: Protein Structure Classification.
Visualize Molecular Surfaces with Protein Workshop
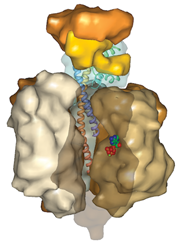
PDB ID 1e79The molecular graphics program Protein Workshop1 can be used to generate molecular surfaces to aid in the display of quaternary structure, protein-protein interactions, and binding sites. This robust feature is able to handle very large assemblies, such as virus capsids.
Surfaces are created for all macromolecule chains in a PDB entry using an algorithm from D. Xu and Y. Zhang.2
A detailed description for using this feature includes: how to create molecular surfaces; color by surface, chain, and entity; making a specific surface transparent; and tips.
Protein Workshop can be launched from every Structure Summary page using the link located underneath the image of the biomacromolecule.
The program can also be used to generate high resolution images for publication.
Web Services for Accessing PDB Data
Web Services can help software developers build tools that efficiently interact with PDB data. Instead of storing coordinate files and related data locally, Web Services let software tools access the RCSB PDB remotely. Detailed documentation for accessing these Web Services is available.
(RESTful | SOAP).
RESTful services exchange XML files in response to URL requests. RESTful search services return a list of IDs for Advanced Search and SMILES-based queries. RESTful fetch services return data when given IDs, including PDB entity descriptions, ligand information, third-party annotations for protein chains, and PDB to UniProtKB mappings. Web Services can also be used to generate pre-defined summary and customizable reports for query results. A new service for accessing pre-released sequences in FASTA format has been recently added.
Improvements are being made based on community feedback. Please let us know if there are website options that you think should be offered as a Web service.
EM Maps Added to PDB Archive
The EM Data Bank (EMDB), the primary archive for experimentally-determined maps obtained using 3D electron microscopy methods, has joined the PDB archive (ftp://ftp.wwpdb.org). With the addition of EMDB data, the physical size of the complete wwPDB archive is now approximately 180 GB. Sites that mirror the full wwPDB archive will need to increase storage capacity accordingly. Summary information regarding the merger and detailed specifications for data access are online at wwpdb.org/em.
- J.L. Moreland, A.Gramada, O.V. Buzko, Q. Zhang and P.E. Bourne (2005) The Molecular Biology Toolkit (MBT): A modular platform for developing molecular visualization applications. BMC Bioinformatics 6:21.
- D. Xu and Y. Zhang. (2009) Generating Triangulated Macromolecular Surfaces by Euclidean Distance Transform. PLoS ONE 4:e8140.
