| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2014 | 269,913 | 635,303 | 1888.15 GB |
| February 2014 | 284,320 | 642,443 | 1897.20 GB |
| March 2014 | 315,871 | 743,752 | 2151.62 GB |
| April 2014 | 291,940 | 689,037 | 1888.15 GB |
| May 2014 | 282,270 | 674,707 | 2165.87 GB |
| June 2014 | 244,617 | 601,935 | 1727.70 GB |
| July 2014 | 218,350 | 599,228 | 1502.77 GB |
| August 2014 | 216,099 | 512,845 | 1525.36 GB |
| September 2014 | 292,187 | 673,001 | 2027.27 GB |
| October 2014 | 345,880 | 795,099 | 2537.37 GB |
| November 2014 | 342,615 | 778,448 | 2981.94 GB |
| December 2014 | 296,236 | 674,383 | 2591.72 GB |
In December 2014, large structures (containing >62 chains and/or 99999 ATOM lines) represented as single files were fully integrated into the main PDB FTP archive in both PDBx/mmCIF and PDBML formats. Previously, large structures were represented in multiple "SPLIT" entries, which have now been removed (obsoleted). Users searching for ID codes of "SPLIT" entries at wwPDB member websites will be automatically redirected to the combined entry.
A separate directory in the PDB FTP archive contains a TAR file including a collection of "best-effort," minimal, PDB format files for large structures that contain authorship, citation details and coordinate data, and an index file that contains the mapping between the chains present in the large entry and the chains present in the limited PDB-format files. DOIs for large structures will point to these TAR files.
Large structures will only be distributed in the main PDB FTP directory in PDBx/mmCIF and PDBML formats, including biological assembly files. Structures that do not exceed the limitations of the PDB format will continue to be provided as PDB files in the archive for the foreseeable future.
A snapshot of the FTP archive before this change is available at RCSB PDB and PDBj.
Several software libraries developed for the RCSB PDB website are available as open source on GitHub at github.com/rcsb.
Available RCSB PDB projects include molecular viewers (Ligand Explorer, Protein Workshop, Simple Viewer, and Kiosk Viewer), detection of symmetry in quaternary assemblies, and Protein Feature View.
Related projects hosted on GitHub include BioJava, a framework for rapid application development in bioinformatics and Biodalliance, the genome-browser library featured in RCSB PDB's Gene View.
In addition, GitHub hosts several front-end libraries used by RCSB PDB. These include twitter bootstrap and jQuery (including extensions jQuery-UI and jQuery-SVG.
Feel free to join our projects, follow us, or fork us!
The home page has been redesigned with a clean, streamlined look. All information related to high-level topics such as Deposit, Search, or Visualize can be browsed from a single panel to offer a 'one stop shopping' experience. The updated top bar navigation menu system provides quick access to tools and information.
In addition, users can now quickly download references to Mendeley & EndNote from the Structure Summary page.
A detailed description of all new website features is available on the What's New Page.
Membrane proteins in the PDB have been identified and annotated to provide improved searching and reporting options.
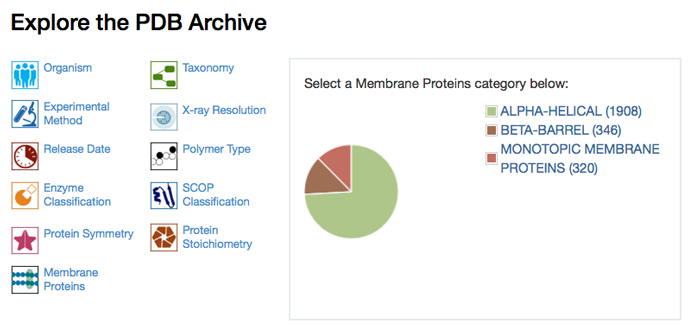
Users can quickly find all annotated membrane proteins in the PDB by entering "membrane proteins" in the top bar simple search and selecting the "Retrieve Membrane Proteins" option.
The Membrane Protein Browser and the Membrane Proteins drill-down tool from the home page and search results can be used to investigate specific membrane protein classifications and access the corresponding structures.
Membrane protein annotations for each entry appear in the Search Results and individual Structure Summary pages (example: 2rh1).
Transmembrane proteins in the PDB are identified using the mpstruc database (Stephen White, UC Irvine), sequence clustering, and data derived from UniProt. Details are available.
Web Services can help software developers build tools that efficiently interact with PDB data. Instead of storing coordinate files and related data locally, Web Services let software tools to access the RCSB PDB remotely. Detailed documentation for accessing these Web Services is available.
RESTful services exchange XML files in response to URL requests. RESTful search services return a list of IDs for Advanced Search and SMILES-based queries. RESTful fetch services return data when given IDs, including PDB entity descriptions, ligand information, third-party annotations for protein chains, and PDB to UniProtKB mappings.
Improvements are being made based on community feedback. Please let us know if there are website options that you think should be offered as a web service.
The RCSB PDB is looking for a Senior Java Web Developer and a Senior Scientist at UC San Diego. Descriptions are available on our Careers page; applications should be submitted online.