| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| July 2015 | 250,325 | 631,670 | 2384.59 GB |
| August 2015 | 245,663 | 593,903 | 2055.11 GB |
| September 2015 | 320,847 | 761,756 | 2874.63 GB |
The Worldwide Protein Data Bank and the Cambridge Crystallographic Data Centre (CCDC) have released a new data resource containing correspondences between the biopolymer components and ligand molecules found in the PDB archive that exactly match small-molecule X-ray structures in the Cambridge Structural Database (CSD) archive.
The chemical structure of every unique molecule in the PDB archive is described in the PDB Chemical Component Dictionary (CCD). The new PDB Chemical Component Model data file complements information in the PDB by providing the following CSD information for matching molecular entries: accession code correspondences, Cartesian coordinates and R-value, data-collection temperature and a disorder flag, SMILES and InChI descriptors, and a Digital Object Identifier (DOI) for the citation associated with the CSD entry.
The CCD contains more than 20,000 components; ~1500 exact match structures have been identified in the CSD. The new PDB Chemical Component Model file is available from the PDB FTP archive. As a service to the scientific community, the wwPDB partners and the CCDC make all these files available freely and without restrictions on use.
Work at the CCDC was supported by BBSRC grant BB/K016970/1 to PDBe and work at RCSB PDB was supported by NSF grant DBI-1338415.

Several software libraries developed for the RCSB PDB website are available as open source on GitHub at github.com/rcsb.
Available RCSB PDB projects include molecular viewers (Ligand Explorer, Protein Workshop, Simple Viewer, and Kiosk Viewer), detection of symmetry in quaternary assemblies, and Protein Feature View.
Related projects hosted on GitHub include BioJava, a framework for rapid application development in bioinformatics and Biodalliance, the genome-browser library featured in RCSB PDB's Gene View.
In addition, GitHub hosts several front-end libraries used by RCSB PDB. These include twitter bootstrap and jQuery (including extensions jQuery-UI and jQuery-SVG.
Feel free to join our projects, follow us, or fork us!
Web Services can help software developers build tools that efficiently interact with PDB data. Instead of storing coordinate files and related data locally, Web Services let software tools to access the RCSB PDB remotely. Detailed documentation for accessing these Web Services is available.
RESTful services exchange XML files in response to URL requests. RESTful search services return a list of IDs for Advanced Search and SMILES-based queries. RESTful fetch services return data when given IDs, including PDB entity descriptions, ligand information, third-party annotations for protein chains, and PDB to UniProtKB mappings.
Improvements are being made based on community feedback. Please let us know if there are website options that you think should be offered as a web service.

Most PDB entries are associated with a primary reference. The option to download citation information for Mendeley and EndNote reference management programs has been added to Structure Summary pages. This citation information is obtained from PubMed, when available.
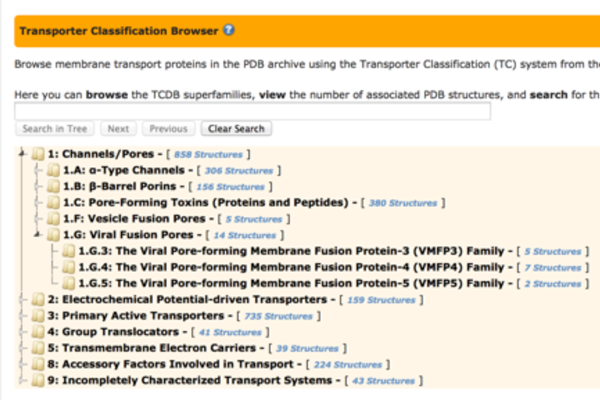
Use the Transporter Classification Browser to find PDB's membrane transport proteins as organized by TC Database family (www.tcdb.org). The browser will autocomplete search terms with the matching classifications, and highlight locations on the tree.
RCSB PDB's Browse Database feature offers access to structures in the PDB archive using different hierarchical trees.
Browsers are available to find structures organized around the following classifications:
Click Browse from the top of every RCSB PDB page and then the desired Browser Tab to start searching.