RCSB PDB's website at rcsb.org was visited each month by an average of 315,538 unique visitors and 741,283 unique visits. A total of 35,260 GB of data were accessed. Traffic is tracked using AWstats.
| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2015 | 306,978 | 708,766 | 2883.29 GB |
| February 2015 | 317,313 | 697,954 | 3299.35 GB |
| March 2015 | 354,060 | 826,464 | 3491.21 GB |
| April 2015 | 330,794 | 777,611 | 3338.75 GB |
| May 2015 | 316,080 | 746,219 | 3262.69 GB |
| June 2015 | 278,575 | 669,676 | 2900.68 GB |
| July 2015 | 250,325 | 631,670 | 2384.59 GB |
| August 2015 | 245,663 | 593,903 | 2055.11 GB |
| September 2015 | 320,847 | 761,756 | 2874.63 GB |
| October 2015 | 378,256 | 896,650 | 3316.23 GB |
| November 2015 | 378,549 | 874,125 | 3187.54 GB |
| December 2015 | 309,023 | 710,610 | 2266.76 GB |
A snapshot of the PDB archive (ftp://ftp.wwpdb.org) as of January 1, 2016 has been added to ftp://snapshots.wwpdb.org/. Snapshots have been archived annually since January 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20160101 includes the 114,679 experimentally-determined coordinate files and related experimental data available at that time. Coordinate data are available in PDB, mmCIF, and XML formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot is 555 GB.
RCSB PDB's top search box helps users easily and intuitively create simple text searches. It recognizes particular types of syntax, such as SMILES strings and sequences, and suggests related search options accordingly.
Typing text in the top search bar launches an interactive pop-up box that suggests possible matches that are organized by categories ranging from author name to ontology terms.
For example, autosuggestions for "bird" will suggest structures with authors whose names contain "bird" and structures from the organism bird. Users who want a simple perform a simple, non-categorized text search can press the "Go" button or keyboard return. Entering SMILES strings will suggest options to perform substructure, exact structure, or similar chemical structure searches, while typing in a sequence will offer different BLAST search options.
Simple text searches complement other RCSB PDB tools: drill down by categorized data distribution summaries, annotation-based browsing using different hierarchical trees, and combining multiple searches of specific data types with Advanced Search.
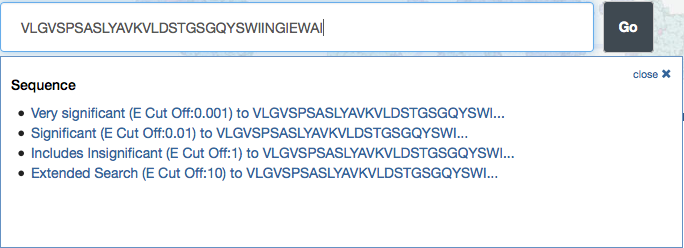
The top menu bar recognizes particular types of syntax, such as SMILES strings and sequences, and suggests related search options accordingly.
Have a list of PDB IDs you'd like to learn more about? Use Advanced Search to view these structures in the Search Results to generate reports and more.
From the Advanced Search pulldown menu, select ID(s) and Keywords > PDB ID(s). Enter the list of multiple IDs, which can be separated by commas or white space, including line breaks.
The structures can then be explored using the different query results browser views: detailed, condensed, timeline or gallery.
Users can also link directly to multiple structures in the Query Results Browser using the syntax:
http://www.rcsb.org/pdb/search/smart.do?smartSearchSubtype_1=StructureIdQuery&structureIdList_1=1MI6+1MVR.
This example will launch entries 1mi6 and 1mvr in the Query Result Browser; other IDs can be added to the URL:
http://www.rcsb.org/pdb/search/smart.do?smartSearchSubtype_1=StructureIdQuery&structureIdList_1=1MI6+1MVR+4GMK+4GSB
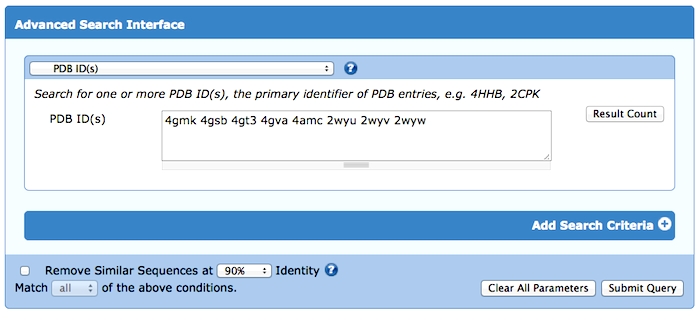
Enter a list of PDB IDs into the Advanced Search.
Protein Feature View graphically summarizes a full-length protein sequence from UniProt and how it corresponds to PDB entries and annotations from external resources in different "tracks".
New tracks illustrate the quality of a protein chain as described in the wwPDB Validation Report and provide a summary of expression tags, cloning artifacts, and other mismatches.
An Overview of Protein Feature View is available, along with examples of the validation and mutation tracks.

A) Protein Feature View of chain A from hemoglobin entry 4HHB from 1984. The PDB Validation track shows several red residues, indicating many geometric outliers. B) Protein Feature View of chain A from hemoglobin entry 2W72 from 1984. The PDB Validation track shows fewer angle and bond outliers, but highlights residues that fit badly to the electron density map (RSRZ >2).
Structure Summary pages have been redesigned to highlight high-level information available for each PDB entry, including structure classification, expression organism, and mutations. More detailed information about macromolecules, small molecules and experimental data and validation appear lower in the page.
These new Structure Summary pages also incorporate "responsive" design that can adjust the layout for mobile devices such as phones and tablets.
As an example, tour the Structure Summary page for the adrenergic receptor PDB ID 2RH1.