
David S. Goodsell holds joint appointments as Associate Professor of Molecular Biology at the Scripps Research Institute and Research Professor at Rutgers University. His research uses computer graphics and simulation to explore structure/function relationships in key biological systems. Current projects include design of compounds to fight drug resistance in HIV and development of methods for computational docking of molecules to proteins and DNA. Science education and outreach is also a strong focus of his laboratory. He is author of the Molecule of the Month, a feature at the RCSB Protein Data Bank that presents the structure and function of a new molecule each month, and several illustrated books on biological molecules, their diverse roles within living cells, and the growing connections between biology and nanotechnology. More information may be found at: mgl.scripps.edu/people/
goodsell
When I was an undergraduate studying biochemistry, one of my teachers presented an enzyme (I forget which one) and described two rival hypotheses for its action. I remember that one of my friends was quite upset about this. We were used to having a list of facts to digest and have ready for exams, and this discussion of hypotheses added unwanted uncertainty to the process. For me, however, it was an exciting moment, when I first saw science as an ongoing process, continually building and growing, with controversies and unknown territory, waiting for me to explore in my own future scientific career.
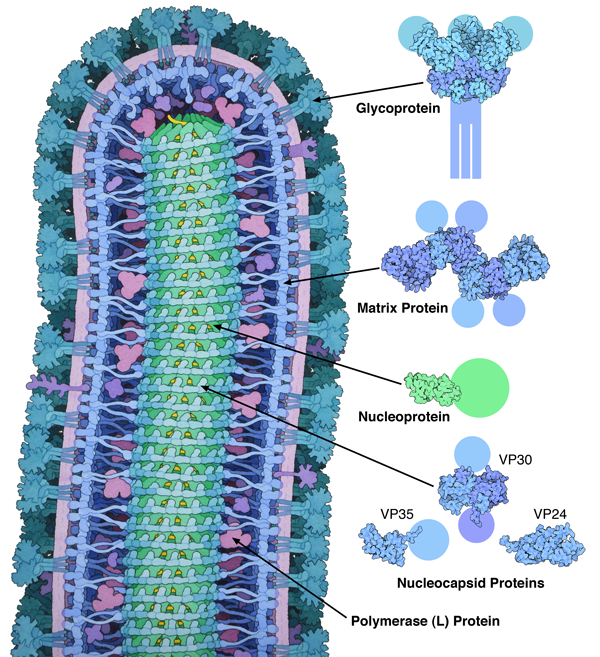
Figure 1. This illustration, which was included in the Molecule of the Month on ebola virus, takes two approaches to representing structural uncertainty. The images on the right are created directly from structures in the PDB, so the images show only what is included in the entry, and the missing parts are represented schematically. The painting of the whole virus at left, however, uses more artistic license in order to present a picture of the whole virus, with no missing parts and drawn in a consistent style. Structures included in the image are from PDB entries 3csy, 4ldd, 4qb0, 3vne, 3fke and 2i8b.
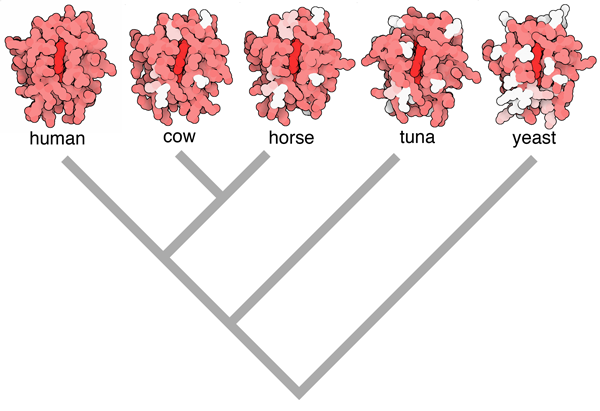
Figure 2. Evolution of cytochrome c can be observed using structures in the PDB archive, by hypothesizing that proteins that are most similar in sequence correspond to organisms that are most closely related. In this illustration, the human enzyme is in red, amino acids that are different in other organisms are colored pink if they are chemically similar, and white if they are completely different. Based on this information, you can build a family tree such as the one shown below the structures. Image from the book Atomic Evidence, using PDB entries 3zcf, 2i8b, 1hrc, 1cyc and 2i8b)
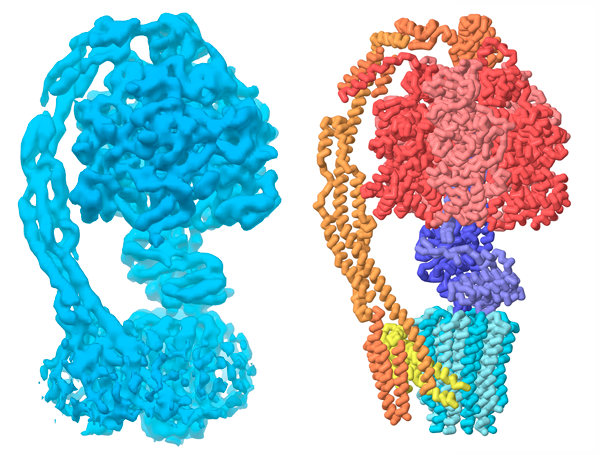
Figure 3. Yeast ATP synthase as seen by cryoelectron microscopy. On the left is the EM map, from entry EMD-4102 at the EMDataBank, and on the right is the atomic structure from PDB entry 5lqz.
As I began doing research, first as an undergrad and then continuing through my graduate work, I got to participate in this search for knowledge. Scientists are continually asking questions and figuring out ways to answer them. The tools we use are very specific in some cases, giving very concrete answers. In other cases, the tools only give us a glimpse of the subject, and we need to combine different methods or infer concepts from the data we have. Sometimes these methods all point to the same answers, other times they give conflicting results and we’re forced to keep exploring to reconcile the observations.
In my work with the Molecule of the Month, I try to capture this intrinsic property of the scientific method: that we understand each subject only to our current state of knowledge. For the most part, structural biology falls strongly in the camp of concrete answers. A crystallographic structure shows the location of atoms in a molecule. Electron microscopy is as close as we can get to a direct look at a molecule. But there are still many aspects that lend uncertainty. Do the (sometimes harsh) conditions of the experiment introduce structural artifacts? Are purified molecules representative of how they act in living cells? Do structures of molecules from one organism provide meaningful information on molecules from a different organism? In each case, we need to weigh the facts and decide if they support our conclusions.
One way I do this is by creating illustrations that only include what is present in the PDB coordinate file. For flexible molecules like P53 tumor suppressor (Molecule of the Month for July 2002), the structures include only the rigid portions of the molecule. For many membrane proteins, such as the RAGE protein presented in June 2015, we may only see the portions on the outside or inside of the cell, which arguably are often the most interesting portions. In these illustrations, I make it clear that we know the structures of some portions, and the rest is shown schematically to show that we’re still working on it.
Sometimes, however, I want to tell a more comprehensive story, so I employ more artistic license. For instance, in the illustration of ebola virus included in the October 2014 Molecule of the Month (Figure 1), I wanted to show the entire virus. I took an integrative approach, gathering as much information as I could find to support the representation of each part of the complex virus. Some parts, like the matrix protein and surface glycoprotein, had been studied by x-ray crystallography, so I could base the shapes of the molecules on atomic information. Other parts, such as the helical nucleocapsid at the center, had been studied by electron microscopy, giving a blurrier view that is still detailed enough to give the overall shape of the assembly. For other portions, such as the polymerase, I could only find sequences and molecular weights, which give an overall size for the molecule.
Recently, I have explored the role of data in our understanding of biology a bit more deeply in a book, “Atomic Evidence: Seeing the Molecular Basis of Life” (Springer 2016). For the book, I picked a collection of central biomolecular concepts, such as molecular evolution (Figure 2) or the central dogma of protein synthesis, and explored the biomolecular structures that support our understanding of the process. Writing this book was a great revelation to me, providing a spur to look at the roots of our scientific knowledge—to see how we know what we know.
In all of this work, on the Molecule of the Month and in the book, I try to promote two overarching ways of thought, both in myself and in readers. The first is: “Get excited.” Excited by the seemingly inexhaustible creativity of scientists and their methods, and excited by the amazing worlds that they reveal. The second is: “Be critical.” We need to search continually for what is known, how it is supported by observation, and how much is still speculation and ripe for additional study.
ATP synthase (Figure 3) is an excellent example of something to get really excited about, but still be critical. Barely a month goes by without somebody asking me to help them find a structure of this amazing molecular machine to use in their classes. However, it’s a tricky subject for structural study, since it has so many moving parts, and until recently, all of the structures in the PDB archive have been various bits and pieces, all separated for study. Recently, researchers have deposited a few structures these rotary motors, such as a yeast F-ATPase (PDB entry 5lqz) and a bacterial V/A-ATPase (PDB entry 5gar), that include the entire assembly. The results are fascinating structures to explore, showing the two connected rotary motors and even capturing them in different rotational states. We need to be critical, though, and realize that these cryoEM structures give us a fuzzy view of the assembly, which was augmented by results from many other studies to trace the path of backbone atoms seen in these PDB entries. So jump in and explore, but think like a scientist and continually ask questions about what you find.