Data Exploration Services
RCSB.org Statistics
Traffic is tracked using internally-developed tools.
| Month | Unique Visitors | Visits |
|---|---|---|
| April 2023 | 725,687 | 3,419,293 |
| May 2023 | 726,558 | 3,832,300 |
| June 2023 | 673,241 | 3,945,158 |
Upload Structure Coordinate Files to Search the PDB
Customize RCSB.org searches by uploading your own experimental data files or Computed Structure Models within seconds. This feature is particularly relevant for AlphaFold2 or ESMfold predictions, homology models, molecular dynamic trajectories, and previously unpublished structures.
In the Advanced Search Structure Similarity and Structure Motif (Figure 1.1) options, use the first drop-down to switch to the “File Upload” mode and click the “Upload File” button to select a local file and upload to RCSB.org (Figure 1.2). Various file formats (.cif, .pdb, .ent) as well as gzipped (.gz) files are supported. Query results can be accessed using a unique URL for 90 days (Figure 1.3). During this time, the link can be shared and/or bookmarked, but is not accessible unless you know its exact URL.

Figure 1. (1) Structure similarity and structure motif search are available from the Advanced Search Query Builder interface. (2) Use the first drop-down to switch to the File Upload mode. (3) Upon file upload the application generates an URL. (4) Use the toggle button to include Computed Structure Models (CSMs) in the search results.
Structure Similarity

RCSB.org uses a novel method to quickly detect similarity between protein shapes of any size.
Use the Advanced Search > Structure Similarity option to find proteins with similar 3D protein shapes using either a PDB ID or a URL to an external structure data file (mmCIF, BinaryCIF or PDB file format). The system attempts to assess global 3D-shape similarity by using BioZernike descriptors that capture the global volumetric shape of the protein. The search will return structures whose volumes are globally similar to the query structure provided.
Structure Motif

Structural motif similarities between PDB structures can provide valuable functional and evolutionary insights.
The Advanced Search > Structural Motif option finds structures containing a small number of specific amino acids in proximity. This feature looks for residues that appear within 15 Å of each other; the residues may be located far apart in the sequence or appear in different polymer chains.
Alternatively, use theMol* standalone 3D viewer to interrogate a local file from your computer. You can then define a structure motif as usual and, upon submission of your query, the structure file will be automatically uploaded.
User Guide Documentation
Detailed instructions are available for using uploaded file for searching bystructural similarity and forrecurring structure motifs.
Toggle to "Opt-in" to Access Computed Structure Models Alongside PDB Data
RCSB.org offers access to >1 million Computed Structure Models (CSMs) from AlphaFoldDB and RoseTTAFold (from ModelArchive).
Only experimentally-determined PDB structures are included in search results by default. Move the "Include CSM" from gray- to cyan-colored slider to activate.

Turn the slider button to cyan to include CSMs when using the Basic Search available at the top of every RCSB.org page or Advanced Search.
Easily Build Advanced Searches
Use Advanced Search > Structure Attributes to query specific fields in the database or to perform a full-text query across all fields.
The database maintains a long list of attribute fields. Users can browse the pull-down menu of options, or search to find relevant categories. For example, type resolution to find the corresponding database attributes that can be queried.
Advanced Search Query Builder can be used combine these Attribute Searches with other types of searches using Boolean operators (AND/OR/NOT):
- Sequence searching: based on a FASTA sequence (with E-value or % Identity cutoffs);
- Sequence Motif searching: find short sequence patterns in PDB structure FASTA sequences using Simple, PROSITE, or RegEx syntax
- Structure Similarity searching: based on an existing Chain or Assembly of a PDB structure
- Chemical Search: find chemical components by Formula or descriptor
Help documentation is available.
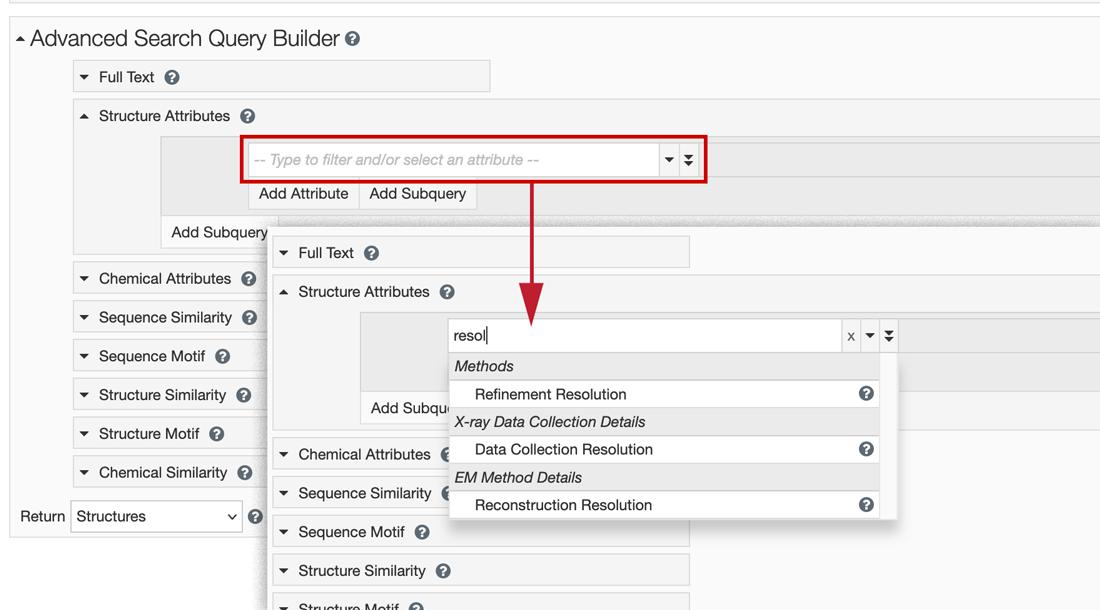
Starting to type the word "resolution" highlights the related attributes that can be searched.
Search for Structures or Feature Help, News, and PDB-101 Articles
Users can target searches towards PDB experimental structures, Computed Structure Models (CSMs), and related data or towards documentation user guides, news, and PDB-101 features.
For 3D biostructure-focused searching, select "3D Structures" from the pulldown option for the RCSB.org search box. This is set as the default. Move the slider to include >1 million CSMs from AlphaFold and ModelArchive in the search.
To search for user guides, news, and PDB-101 content, select the "Documentation" option.

Perform Improved Pairwise Structure Alignments
An improved interface allows for simultaneous analysis and visualization of three-dimensional (3D) structure alignments and structure-based one-dimensional (1D) sequence alignments. The two panels displaying alignment results are interconnected and selecting residues in one panel will select and highlight the same residues in the other panel.
Access the Pairwise Structure Alignment Tool from the Analyze menu at the top of the page or directly from RCSB.org/alignment.
User Guide Documentation is available, with examples of aligning PDB experimental structures with Computed Structure Models (CSMs) from AlphaFold and aligning PDB structures to related enzymes.
Click the image to launch a structural comparison of toxins produced by sea anemones
