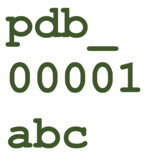Message from RCSB PDB
Training Events
RCSB PDB will be hosting a variety of training events throughout 2024, from virtual Office Hours to specialized Webinars.
Fall Events
Virtual Office Hour: RCSB.org and Advanced Search
Tuesday October 8, 2024
4:00 pm – 5:00pm Eastern |1:00 pm – 2:00pm Pacific
RCSB.org’s Advanced Search feature is a powerful tool for searching the PDB Archive. Want to learn how to better use Advanced Search? Bring questions to our virtual office hour with Rachel Kramer Green, RCSB PDB’s Scientific Support & Customer Service Lead.
Registration is required for the Zoom meeting information, but attendance is at no charge. Please sign up at https://go.rutgers.edu/2ueche6f.
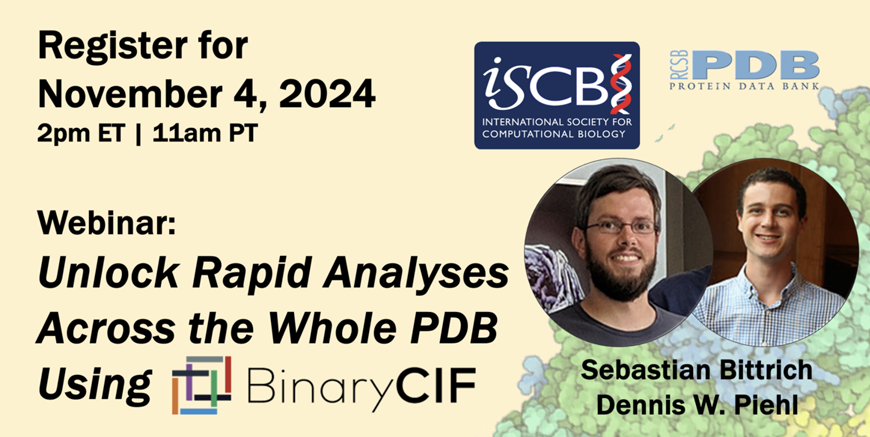
Webinar:
Unlock Rapid Analyses Across the Whole PDB Using BinaryCIF
Monday November 4, 2024
2:00 pm – 3:00 pm Eastern | 11:00 am – 12:00 pm Pacific
Join our one-hour workshop to future-proof your data analysis with BinaryCIF, a fully interchangeable yet drastically more efficient flavor of the PDBx/mmCIF format. BinaryCIF not only boosts storage efficiency, but also substantially improves parsing speed, making it ideal for large-scale analyses. BinaryCIF is supported by resources such as RCSB PDB, PDBe, and AlphaFold DB.
This webinar will benefit bioinformaticians, data scientists, and structural biologists who want to
- Understand the basics of the PDBx/mmCIF schema
- Access BinaryCIF files and related APIs on RCSB.org
- Programmatically consume BinaryCIF data and convert between formats
- Compute archive-wide statistics across the entire PDB
- Gain hands-on experience with our Python parser
This webinar is part of the ISCBacademy series.
Registration is required for the Zoom meeting information, but attendance is at no charge. Please sign up at https://go.rutgers.edu/y84mir74.
Office Hour:
Supporting Extended PDB IDs
Thursday, November 7, 2024
2:00 pm – 3:00 pm Eastern | 11:00 am –12:00 pm Pacific
The wwPDB anticipates that all four-character PDB IDs will be exhausted by 2028, after which 12-character PDB IDs will be issued. Entries with extended PDB IDs will not be compatible with the legacy PDB file format and will only be available in PDBx/mmCIF format. wwPDB encourages users to transition to the PDBx/mmCIF format as soon as possible.
Bring any related questions to our virtual office hour with RCSB PDB's Ezra Peisach.
Registration is required for the Zoom meeting information, but attendance is at no charge. Please sign up at https://go.rutgers.edu/p0u1dhuq.
Software developers who will need to make updates to code are encouraged to attend.
Past events
The recording and related materials from our Summer events Python Scripting for Molecular Docking and Teaching Enzymology with the PDB: From Pandemic to Paxlovid are available from PDB-101.
Sign up for our Training Announcements mailing list or contact info@rcsb.org for more information.
| Snapshot: October 1, 2024 | |
|---|---|
| 225,399 | Released atomic coordinate entries |
| Molecule Type | |
| 194,767 | Proteins, peptides, and viruses |
| 4,526 | Nucleic acids |
| 13,610 | Protein/nucleic acid complexes |
| 12,261 | Protein/Oligosaccharide |
| 22 | Oligosaccharide (only) |
| 213 | Other |
| Experimental Technique | |
| 188,017 | X-ray |
| 14,381 | NMR |
| 22,644 | Electron Microscopy |
| 236 | Multi Method |
| 82 | Neutron Diffraction |
| 37 | Other |