Education Corner
Molecule of the Month: Celebrating 25 Years of Storytelling and Announcing New Beginnings
After a remarkable 300 columns, David Goodsell has retired from the Molecule of the Month series. Janet Iwasa will be continuing the series for PDB-101, beginning with the January 2025 article on Assembly Line Polyketide Synthases
The PDB and Me

David S. Goodsell
Scripps Research and Rutgers University
The Protein Data Bank has been a constant companion throughout my career, providing a place for me to share my work and a place to visit for inspiration. I began my career as an X-ray crystallographer in the laboratory of Richard Dickerson at UCLA, determining the structures of small DNA oligonucleotides and their complexes with toxic, and potentially therapeutic, small molecules. My first experiences with the PDB was as a depositor, working with curators to produce clean, annotated versions of my structural entries. To this day, I remember the excitement of seeing my first electron density map and actually being able to see first hand the phosphates and bases in the DNA double helix that I had synthesized--and the pride of being able to share the atomic coordinates in the PDB archive.
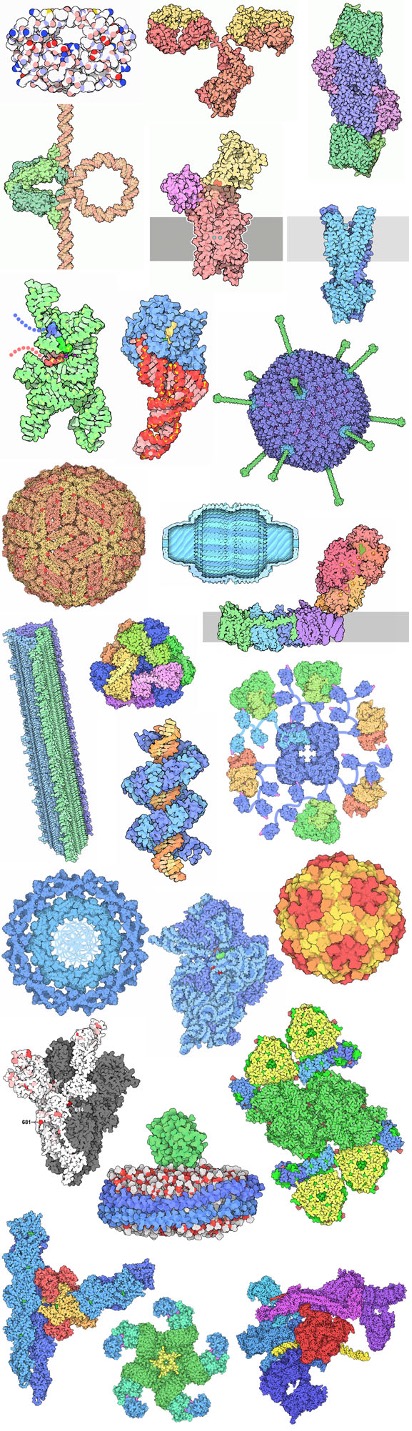
Selections from 25 years of the Molecule of the Month.
I also quickly became a voracious consumer of PDB files as I worked to make sense of the structures I was determining. At the time, the laboratory was interested in exploring the molecular mechanics of DNA bending, to help understand how DNA interacts with drugs and proteins. The PDB allowed us to access structures from around the world, to develop hypotheses that reconciled the local nucleotide geometries across them all. Unfortunately, there was a fierce battle being waged in the structure community about deposition of coordinates--some laboratories chose to keep their coordinates secret. Being an annoying student, I peppered these laboratories with requests, and if I was turned down, I extracted coordinates from stereo pairs printed in their publications. Fortunately, the culture of structural biology changed soon after as funding agencies and journals added requirements for coordinate deposition.
I have had the pleasure of watching the PDB grow over the course of my career. When I started graduate school, the PDB included a grand total of 117 structures and it was easy to browse through every one individually. My first PDB entry, a structure of DNA bound to netropsin determined with Mary Kopka and others in the Dickerson lab, was released a few years later as the 175th structure released in the archive. In my postdoctoral work with Arthur Olson at the Scripps Research Institute, I took a more active role in watching the PDB grow, surveying the entire archive to explore topics such as the role of protein symmetry in protein function and the detailed atomic characteristics of protein-protein interfaces.
When I started work in my own laboratory, I had the opportunity to play a more active role at the PDB. I was writing short molecular stories to test a new interactive molecular viewer, and Helen Berman offered a place for them on the RCSB PDB website to help with outreach. These stories soon became the Molecule of the Month. When we had built up a suitable collection of articles, Helen brought together the entire staff of the PDB and we brainstormed about how to leverage the collection for education and outreach, which led to the creation of PDB-101. Later, when Stephen Burley took the reins, he spearheaded additional efforts to expand the audience of PDB-101 and provide targeted materials related to current global health issues.
The Molecule of the Month has been a source of pleasure throughout my career, allowing me the opportunity to explore a wide range of fascinating topics. I’ve witnessed many revolutions in structural science, including the mapping of protein folding space with structural genomics and its use in protein structure prediction, time-resolved crystallography to see enzymes in action, and most recently, the use of cryoelectron microscopy to explore assemblies of unprecedented complexity. I’ve watched as structures got ever larger, and ever more detailed in both time and space. And throughout this, the free, open access policy of the PDB has allowed me the freedom to create pictures of anything that caught my fancy.
I’m very happy to announce that I will be passing the baton with my 300th installment of the Molecule of the Month. Janet Iwasa, a world leader in biomolecular animation and outreach, will be continuing the project. I’m very much looking forward to seeing how she brings her unique artistry to PDB-101!
Milestone Molecules of the Month
- Adrenergic Receptors: 100th column (2008)
- PDB Pioneers: celebrating PDB’s 40th anniversary (2011)
- Fifty Years of Open Access to PDB Structures: celebrating PDB’s 50th anniversary (2021)
- Quasisymmetry in Icosahedral Viruses: 200th column (2016)
- All Molecule of the Month articles by Date
Continuing a Visual Legacy

Janet Iwasa
The University of Utah and RCSB PDB
I am honored and excited to be taking on the Molecule of the Month from David Goodsell. But, if we're being honest, I should admit that when David first asked me about taking on this responsibility, I felt apprehensive. As a long-time follower and fan of PDB-101, I am aware of the huge impact David's Molecule of the Month articles have had on diverse communities. His enormous shoes just seem impossible to fill.
David has long been one of my heroes; he was one of the first people I contacted when I was first considering a career in molecular visualization during graduate school, and I have since benefited from David's thoughtful advice as a colleague and collaborator. David's beautiful illustrations and the careful consideration he has put into the visual representation of the molecular world have also heavily influenced my own work in molecular animation.
I am happy to accept the baton passed to me by David, and grateful for this opportunity to continue David's wonderful legacy. I look forward to continuing the tradition of sharing interesting and accessible molecular stories describing the structures of the PDB, and visualizing these concepts through colorfully rendered illustrations and, yes, animations (Check out January's Molecule of the Month article to see an animation of an assembly line polyketide synthase)!