|
|
|
|
|
||
RCSB PDB Focus: Sorting Search Results and Tabular Reports Query results can be organized in an image gallery  A search results list sorted by Release Date  Citation report for a search results set. The RCSB PDB offers many ways of looking at the information contained in the database. After searching for a set of structures, users can explore individual structures or examine the whole set by creating reports.
Following a search that produces multiple entries, the results set can be sorted by choosing 'Sort Results' from the menu on the left hand side of the page. Sorting options include: PDB ID, Release Date, Residue Count, Resolution and Rank. An Advanced Search by sequence (Advanced Search>>Sequence Features>>Sequence (Blast/Fasta)) allows the user to sort results by PDB ID, formula weight and E value.
Another option for viewing multiple structure results is available from the left menu's "Tabulate" button. It allows the user to create tables of various structural and experimental properties that can be downloaded as CSV files.
These tables can be sorted by clicking on the column headers. Clicking again reverses the sort order.
The "Custom Report" option lets the user select which columns will be included in the report. "Collage" will tile the thumbnails of all of the molecular images.
Default reports about structure, sequence, ligands, primary citation, and biological details are available from the Summary Reports option. Experimental reports can also be created for X-ray (crystallization, data collection, refinement, refinement parameters, unit cell) and NMR (representative model, spectrometer, sample conditions, software, refinement, and ensemble) structures.
RCSB PDB Focus: Exploring Domains in Protein Structure Display options on the Sequence Details page Domains can be thought of as the smallest structural units from which proteins are assembled that retain properties of the whole protein, such as a hydrophobic core. In certain cases, domains can also function independently from the rest of the structure. Any given protein structure is comprised of one or more domains from which the overall properties of the protein are derived. Analyzing a protein structure from the point of view of its composite domains is an important, yet not fully solved problem. The RCSB PDB offers various ways of exploring domains in protein structures: PDOMAINS (http://pdomains.rcsb.org/pdomains/)1 is a resource centered around the definition and assignment of structural domains in proteins. It offers analysis of existing approaches to domain definition and provides a benchmark dataset to evaluate and cross-compare automatic domain assignment methods. The Browse Database option of the 'Search' tab on the left-hand menu offers a tool to explore the hierarchically organized and curated domain definitions produced by SCOP and CATH.  SCOP Browser Structure Summary pages for individual structures provide links to sets of all structures containing domains similarly categorized by SCOP and CATH.  SCOP and CATH data on the Structure Summary page. Each annotation is a link to a set of PDB structures. Sequence Details pages for each structure illustrate domains aligned with sequence and secondary structure. The colored domain definition links in the SCOP Domains section coincide with the colored bars underneath the sequence to indicate the location of the domains. Boundaries between domains are highlighted further with vertical dashed lines.  Sequence Details page The user can choose domain definitions according to either SCOP or CATH and the programs DomainParser and PDP.  Display options on the Sequence Details page 1Holland TA, Veretnik S, Shindyalov IN, Bourne PE. Partitioning protein structures into domains: why is it so difficult? J Mol Biol. 2006 Aug 18;361(3):562-90.
Searching for Sequence VariantsProtein structure sequences are assigned UniProt/SwissProt IDs(UNP/SWS). The new Sequence Variants/Non-variants RCSB PDB feature lets users retrieve all structures with a particular SWS/UNP ID, grouped by the presence or absence of sequence variations (variants or non-variants). Searches for variants will provide structures with post-translational modifications, whereas searching for non-variants will provide occurrences of structures that have at least one identical polypeptide chain. This query can be run using the Advanced Search option.
 The Advanced Search form for Sequence Variants Variants and non-variants can also be retrieved from the Structure Summary page for any structure having such variants with the same UNP/SWS ID. From the lefthand menu, select Structure Analysis-> Sequence Variants. The resulting page displays the pairwise alignment of the structure sequence and the UNP/SWS sequence for each structure.  Structure Summary pages offer options for viewing Sequence Variants from the "Structure Analysis" option
 Variations are color coded in the alignment display
RCSB PDB Focus: External LinksStructure Summary pages for all structures in the PDB now offer a set of external links. Accessible from the left menu, these links provide further information about the structure under study, such as biochemical pathway information, stereochemistry and ligand binding data. 
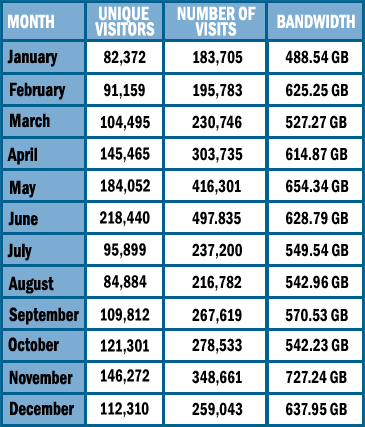
WEBSITE STATISTICSAccess statistics for www.pdb.org are given below for the year 2006.
|
||
©2007 RCSB PDB |