for Structural Bioinformatics Protein Data Bank
Fall 2008
Number 39
| EDUCATION
CORNER
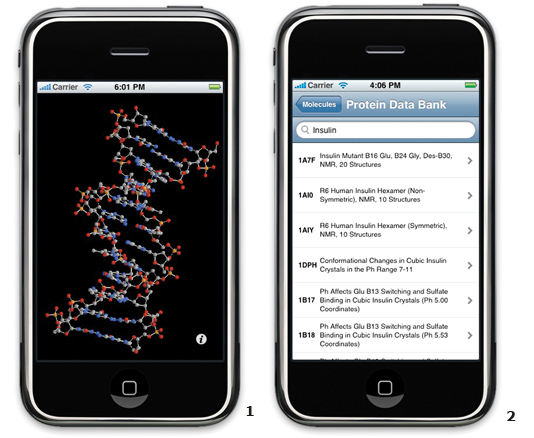
In recent years, we’ve seen an explosion in processing power within portable devices such as cell phones and media players. Current devices more closely resemble general-purpose computers, especially Apple Inc.’s new iPhone and iPod Touch. Both use a form of the OS X operating system found on Macintosh computers, and can run custom applications designed using a desktop-caliber software development kit (SDK). Additionally, these applications are available worldwide through Apple’s iTunes Store and can be downloaded with a single click. Many high school and college students will carry these devices into classes this fall, providing a opportunity for educational software to be used in unique ways. Molecules is one such application that will hopefully introduce students to the fascinating world of biomolecules and their 3D structures. One of the inspirations for Molecules was a conversation I had with my brother, Matt Larson, who is using X-ray crystallography to determine the structure of a protein as part of his Ph.D. work at the University of Alabama at Birmingham. He was preparing to present a poster with an early structure for this protein, and I couldn’t help but think that a static image wasn’t the best means of displaying that structure. After witnessing the capabilities of the iPhone and iPod Touch, I imagined what it would be like if he could take an iPod to a conference and pull it out of his pocket every time he wanted to show off the full 3D structure of his protein or even download and research on the spot any PDB molecule mentioned in a presentation. Molecules for the iPhone/iPod Touch: Molecules is a free, open-source molecular visualizer that uses the built-in 3D hardware acceleration, touch interface, and always-on data networking capability of the iPhone to deliver an intuitive means of viewing molecular models away from a desktop computer. (I will refer to the iPhone for the remainder of the article, but the iPod Touch shares all of the same hardware, minus the cell phone connection, and thus can also run Molecules). Molecules allows users to look up and download files from the PDB to the iPhone, even over the cell phone data network. Once the files are on the device, molecules currently can be displayed in ball-and-stick, spacefilling, and cylinder (colored by amino acid or nucleotide residue) visualization modes. To rotate the models, users simply move their finger across the display. A molecule can be zoomed out by pinching two fingers together or zoomed in on by reversing the gesture. Panning across the molecule is accomplished by dragging it using two fingers. Using Molecules in the Classroom
1. B-DNAa rendered in the ball-and-stick visualization mode in Molecules Initial response: Because of reasonably fast rendering performance, these easy-to-learn controls provide the impression that you are manipulating the molecule itself with your fingers. The response I’ve heard over and over from non-scientific users who have tried this out is “That’s so cool!” This is exactly what you’d like to hear from students, especially impressionable younger ones. What they have viewed as just their cell phone or music player suddenly becomes a scientific tool with the addition of one program. This capability, coupled with a savvy educator, could make a significant impact. This is not just wishful thinking on my part. I’ve received correspondence from professors and high school teachers who will be using this as an aid in their upcoming classes. I’ve also heard from many researchers across the world who are very excited about the prospects of using this as an extension of their desktop software, and as a tool that allows them to look up structures while out in the lab or at a conference. Most surprising to me has been the number of people who have used Molecules. In the first three months of availability, it has been downloaded by more than 230,000 unique users. Some of its popularity comes from being a free iPhone application, among the first 500 released, and from being listed as a Staff Favorite on the front page of iTunes. Still, that is a rather large number of users for a niche application. Future plans: Along with all the positive reviews and messages have come some complaints. The first was about how difficult it was to find and load new structures onto the iPhone. Initially, I required users to type in the four-character ID code to download new structures. Even with an integrated web browser to search within the RCSB PDB website, many users found this frustrating. With the help of the RCSB PDB, I was able to use Web Services to integrate keyword searching into Molecules itself. Web Services allow you to communicate directly with the RCSB PDB’s data store and retrieve values without having to go through the website; this is ideal for a portable device on a bandwidth-limited connection. Another area where Molecules falls short is the amount of time it takes to load large structure files, render them to the screen, and refresh the display after changes are made in orientation or scale. This makes working with structures containing over 10,000 atoms a chore. While you might think that this is a natural limitation of the portable hardware, the truth is that the iPhone has a surprisingly fast processor and graphics chip, and I believe it is capable of much more. I am currently testing a series of enhancements that should make file loading and initial rendering much faster, and am learning from other open source projects such as QuteMol (qutemol.sourceforge.net) that appear to have far more efficient 3D engines than Molecules. Finally, other reviewers have identified two additional requirements to make this more useful as a scientific tool: additional visualization modes, primarily ribbon representations, and the ability to load custom structures not contained within the PDB. Both of these enhancements will take a little work, but I would expect to accomplish them within the next few months. One pleasant result of the initial exposure of this program has been the offers of help from other developers, some experienced in writing other visualization applications. Their assistance should speed along the implementation of ribbon and other display modes. Loading custom molecules onto the device presents a tricky problem, given some of the limitations of the mobile platform, but other iPhone programmers have suggested a solution. I will be crafting a desktop version of this application that can load whatever custom structures you would like and store them locally in a manner similar to iTunes. A desktop client would then be able to synchronize to a users’ iPhone through a network connection to provide iPhone access to whatever structures are on the computer. Conclusion: When it came to choosing data sources to use for Molecules, the RCSB PDB was first on my list. I have been a fan of the work they have done for years. I’d like to thank them again for providing this resource and being so helpful in the development of this program. This whole experience has been a lot of fun and hopefully will result in an application that students and researchers alike will find useful. I am excited to see what other scientific applications will emerge for portable devices, once people realize that these are no longer just pieces of consumer electronics but powerful mobile computers. For more information: Blog posts and tips on programming the iPhone’s 3D hardware are at www.sunsetlakesoftware.com/molecules. A direct link to the free Molecules download from iTunes is provided for users with a compatible device (iPhone or iPod Touch with the 2.0 firmware update installed). To participate in a discussion or read what others have had to say, stop by the forums at www.sunsetlakesoftware.com/forum. Finally, the source code to Molecules has been released under the BSD license at the first page mentioned above. In addition to the educational opportunities of the application itself, I hope that the availability of the code will help others develop similar portable scientific tools. a1bna: H.R. Drew, R.M. Wing, T. Takano, C. Broka, S. Tanaka, K. Itakura, R.E. Dickerson (1981) Structure of a B-DNA dodecamer: conformation and dynamics. Proc.Natl.Acad.Sci.USA 78: 2179-2183. |
E-mail: info@rcsb.org • Web: www.pdb.org • FTP: ftp.wwpdb.org
The RCSB PDB is a member of the wwPDB (www.wwpdb.org)