Data Deposition and Annotation
Depositing Structures with Ligands
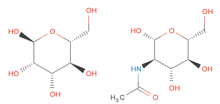 Ligand Expo can also be used to check and confirm the chemistry of a chemical component. Shown is the correct sterochemistry for alpha-D-mannose (on the left, ID: MAN) and for N-acetyl-D-glucosamine (on the right, ID: NAG).
Ligand Expo can also be used to check and confirm the chemistry of a chemical component. Shown is the correct sterochemistry for alpha-D-mannose (on the left, ID: MAN) and for N-acetyl-D-glucosamine (on the right, ID: NAG).
Ligand Expo (ligand-expo.rcsb.org) accesses chemical and structural information about all small molecule components found in PDB entries. It is based upon the Chemical Component Dictionary maintained by the wwPDB. When depositing a structure with a ligand:
- Search Ligand Expo for a chemical component that matches your ligand
- If a match is found, use the corresponding three-character code for the ligand in your coordinates
- If the ligand is new, choose a new three-character code for the ligand
- When depositing your structure with ADIT, upload the chemical name and formula and/or a file showing the chemical image for the new ligand into the Ligand Information section
Deposition Statistics
From April 1-June 30, 2012, 2506 experimentally-determined structures were
deposited to the PDB archive, and then processed and annotated by wwPDB teams.
Of the 4978 structures deposited in the first half of 2012 (January 1-June 30),
80% were deposited with a release status of "hold until publication"; 15%
were released as soon as annotation of the entry was complete; and 5% were
held until a particular date. 93% of these entries were determined by X-ray
crystallographic methods; 6% were determined by NMR methods.
During the same time period, 4520 structures were released and made publicly
available in the PDB.
wwPDB News:
Journal of Biological Chemistry to Require Validation Reports
for Structure Papers
By the end of Summer 2012, The Journal of Biological Chemistry (JBC) will require authors to submit a validation summary report along with manuscripts describing X-ray crystallographic structure studies.
Validation reports are already required by the International Union of Crystallography (IUCr) journals as part of their submission process.
wwPDB members currently provide depositors with detailed reports that include the results of geometric and experimental data checking as part of the structure annotation process. These documents are available from all wwPDB annotation sites as PDF files for easy sharing and review.
The validation reports will continue to be developed and improved as we receive recommendations from our Validation Task Forces for X-ray,1 NMR, EM,2 and small angle scattering methods, and as we further develop our data deposition and processing procedures and the wwPDB Common Deposition & Annotation Tool.
More information about JBC's announcement can be found at ASBMB Today
- R. J. Read, P. D. Adams, W. B. Arendall, III, A. T. Brunger, P. Emsley, R. P. Joosten, G. J. Kleywegt, E. B. Krissinel, T. Lutteke, Z. Otwinowski, A. Perrakis, J. S. Richardson, W. H. Sheffler, J. L. Smith, I. J. Tickle, G. Vriend, P. H. Zwart (2011) A new generation of crystallographic validation tools for the Protein Data Bank. Structure 19: 1395-1412.
- R. Henderson, A. Sali, M. L. Baker, B. Carragher, B. Devkota, K. H. Downing, E. H. Egelman, Z. Feng, J. Frank, N. Grigorieff, W. Jiang, S. J. Ludtke, O. Medalia, P. A. Penczek, P. B. Rosenthal, M. G. Rossmann, M. F. Schmid, G. F. Schroder, A. C. Steven, D. L. Stokes, J. D. Westbrook, W. Wriggers, H. Yang, J. Young, H. M. Berman, W. Chiu, G. J. Kleywegt, C. L. Lawson (2012) Outcome of the first electron microscopy validation task force meeting. Structure 20: 205-214.
