Outreach and Education
Join the RCSB PDB Team
We're looking for a Lead Web Architect. Learn more.
2012 Annual Report
The recently published Annual Report highlights the RCSB PDB's accomplishments for 2012. It also reviews topics ranging from deposition statistics to domain-based structural alignments to popular help desk questions and answers.
wwPDB efforts, including the development of the Common Deposition and Annotation Tool, are also highlighted.
This edition's cover maps the proteins and chemical reactions involved with the citric acid cycle.
These bulletins provide a yearly snapshot of RCSB PDB activities and the state of the PDB archive. This edition, the RCSB PDB's thirteenth, is available as a PDF download from www.rcsb.org/ar12. If you would like a printed copy, please send your postal address to info@rcsb.org.
Papers Published
- The RCSB Protein Data Bank: new resources for research and education (2013) Nucleic Acids Research doi:10.1093/nar/gks1200
Update of new tools and resources that provide a structural view of biology for research and education, including improved methods for simple and complex searches of PDB data, creating specialized access to chemical component data and providing domain-based structural alignments. - Creating a community resource for protein science (2012) Protein Science 21: 1587-1596 10.1002/pro.2154
Discusses how management, data content, infrastructure, and community have influenced the continued development of the PDB. It also analyzes the corpus of data to identify trends in the field of structural biology. - BioJava: an open-source framework for bioinformatics in 2012 (2012) Bioinformatics 28: 2693-2695 doi:10.1093/bioinformatics/bts494
Describes modules that provide state-of-the-art tools for protein structure comparison, pairwise and multiple sequence alignments, working with DNA and protein sequences, analysis of amino acid properties, detection of protein modifications and prediction of disordered regions in proteins as well as parsers for common file formats using a biologically meaningful data model. - Illustrating the Machinery of Life: Viruses (2012) Biochemistry and
Molecular Biology Education 40: 291-296 doi:10.1002/bmb.20636
David Goodsell's series continues with illustrations of poliovirus, influenza, and HIV. - Putting proteins in context (2012) Bioessays 34:718-20 doi:10.1002/bies.201200072
David Goodsell describes his thoughts and process for scientific illustrations.
Explore the Structural Biology of HIV
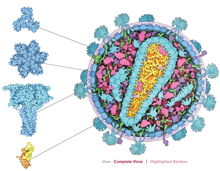
Visit PDB-101 for this animation
and more
Together, these molecules allow the virus to infect cells of the immune system and force them to build new copies of the virus. Each molecule in the virus plays a role in this process, from the first steps of viral attachment to the final process of budding.
The structural proteins, viral enzymes, and accessory proteins of HIV that have been determined are highlighted in a downloadable poster and an interactive, online Flash website. Clicking on a protein in the animation reveals information about the structure and a link to the PDB entry.
The animation and poster are available from PDB-101.
Educational Activities and Lessons
The PDB-101 website packages together resources that promote exploration in the world of proteins and nucleic acids for teachers, students, and the general public.
The Educational Resources section of PDB-101 offers different materials to help learn about structures in the PDB, including posters, animations, and classroom activities and lessons.
These activities and lessons include:
- Exploring the Structure of tRNA. Build a paper model of the primary, secondary, and beginnings of the tertiary structure of tRNA. Visit the related activity page to learn more. Virus Structure Lecture, Lesson Plans, and Activities. These resources, aimed at middle and high school classrooms, include templates to make 3D paper models of disease-causing viruses and review questions.
- Green Fluorescent Protein Tutorial. With this tutorial you will find protein structures using search tools on the RCSB PDB website; use molecular visualization tools to explore the GFP structure and function; and find the GFP gene and view important mutations. 3D Paper Model of DNA. Download a PDF to build one complete turn of a DNA double helix, with the major and minor grooves labeled.
- Education Corner. Other activities and lessons are available from the RCSB PDB Newsletter's Education Corner archives.
Turn Your Computer into a PDB Structure Kiosk
Highlight structures from your lab, institution, or class with the Molecules in Motion Kiosk Viewer. Using a list of PDB IDs, this full-screen animation program will display any structure from different angles and perspectives. It also focuses on any chemical components within the structure. The Java viewer can be downloaded or launched from PDB-101's Educational Resources page.