In the fourth quarter of 2013, 2450 experimentally-determined structures were deposited to the PDB archive for a total of 10561 entries deposited in the year. 9972 entries were deposited in 2012.
Of all structures deposited in 2013, 83.7% were deposited with a release status of hold until publication; 13.7% were released as soon as annotation of the entry was complete; and 2.6% were held until a particular date. 91.8% of these entries were determined by X-ray crystallographic methods; 5.6% were determined by NMR methods.
9630 structures were released in the PDB archive in 2013. They account for ~10% of the current total holdings of 96696. The PDB archive is on course to reach 100,000 entries in 2014: The International Year of Crystallography.
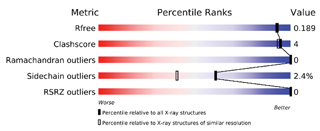
The validation reports provide at-a-glance summary
information
that compares the quality of a model
with that
of other models in the archive.1,2
X-ray structure validation reports can now be generated on demand by macromolecular crystallographers by using the new stand-alone wwPDB validation server.
The new reports implement recommendations of a large group of community experts on validation.1,2 They can be used to assess the quality of early, intermediate and near-final models to identify any potential problems that need addressing prior to structure analysis, publication and deposition.
The stand-alone validation server was developed in the context of a larger wwPDB collaboration, the new wwPDB Deposition and Annotation System, which was created to unify the annotation tools and practices used across all wwPDB deposition centers. The new deposition system, currently in testing, supports all experimental methods currently archived by the wwPDB. The new system incorporates tools for validation of deposited structures as well as all of the chemical, sequence, structure and administrative annotation tasks.
The wwPDB has established PDBx/mmCIF as the new standard format for data exchange and archiving in structural biology.
To help facilitate the transition from PDB to PDBx/mmCIF format, the wwPDB has hosted programmer's workshops that described the content and organization of PDBx/mmCIF data, and the available software tools and libraries supporting PDBx/mmCIF (C/C++, Java, and Python).
The Workshop on the PDBx/mmCIF Data Exchange Format for Structural Biology was held on October 22, 2013 at Rutgers, The State University of New Jersey in conjunction with the PSI Workshop on Theoretical Model Archiving and Validation. This workshop included presentations from: Paul Adams (Phenix), David Case (AMBER), Tom Goddard (UCSF Chimera), Robert Hanson (JMol), Eugene Krissinel (CCP4/mmdb), Andreas Prlic (BioJava), John Westbrook (RCSB PDB). Attendees also worked hands-on with the presenters and developers in the areas of structural biology and molecular modeling to learn application details and to discuss successful approaches and experiences in adapting software to support PDBx/mmCIF.
Another Workshop on mmCIF/PDBx for Programmers took place November 20-21, 2013 at EMBL-EBI in Cambridge, United Kingdom. This event hosted lectures by experts in mmCIF/PDBx and developers of language-specific libraries or packages (C/C++, Java, Python), tutorials, and individual "code hacking". Presenters included Paul Adams, Eugene Krissinel, Garib Murshudov (Refmac), Andreas Prlic, Sameer Velankar (PDBe) and John Westbrook. This workshop followed the Workshop on a Unified Format for NMR Restraints Data (November 18-19, also held at EMBL-EBI).
Additional information about PDBx/mmCIF resources is hosted at http://mmcif.wwpdb.org.
H. Berman, G. J. Kleywegt, H. Nakamura and J. L. Markley. (2013) Comment on Timely deposition of macromolecular structures is necessary for peer review by Joosten et al. (2013) Acta Cryst. D69: 2296.
H. Berman, G. J. Kleywegt, H. Nakamura and J. L. Markley (2013) Comment on On the propagation of errors by Jaskolski (2013) Acta Cryst. D69: 2297.
J.Y. Young, Z. Feng, D. Dimitropoulos, R. Sala, J. Westbrook, M. Zhuravleva, C. Shao, M. Quesada, E. Peisach, H.M. Berman. (2013) Chemical annotation of small and peptide-like molecules at the Protein Data Bank. Database 2013: bat079.