
Rebecca F. Alford received her B.S. in Chemistry from Carnegie Mellon University in 2016 and is currently a Ph.D. student in Chemical and Biomolecular Engineering at Johns Hopkins University under the mentorship of Dr. Jeffrey J. Gray. Rebecca started research as a high school student by combining biomolecular modeling and machine learning to decipher the connection between mutations, biomolecules, and disease. Her Ph.D. research focuses on the thermodynamics underlying biomolecular modeling techniques, specifically for modeling proteins embedded in cell membranes. She built an object-oriented framework for membrane protein prediction and design. Rebecca was honored with the Service to Rosetta Award for her work to increase diversity in the Rosetta biomolecular modeling community. She was a co-instructor at both Rosetta intern boot camps. Rebecca is a Hertz Foundation Fellow and a National Science Foundation Graduate Research Fellow.
Links: @rebeccaalford

Jeffrey J. Gray received his B.S.E. in chemical engineering at the University of Michigan and his Ph.D. in chemical engineering at the University of Texas at Austin. He completed post-doctoral training researching protein-protein docking at the University of Washington. In 2002 he joined the Department of Chemical and Biomolecular Engineering at the Johns Hopkins University in Baltimore, Maryland where he is now a Professor. Gray’s research focuses on computational protein structure prediction and design, particularly protein-protein docking, therapeutic antibodies, and protein-surface interactions. Gray’s lab leads the development of RosettaDock, RosettaAntibody, the ROSIE web server, and the PyRosetta interactive platform for protein structure prediction and design. Gray’s lab has produced the most accurate complex structure for several targets in the CAPRI blind protein-protein docking challenge and sub-angstrom antibody binding loop structures in the Antibody Modeling Assessment. Gray has received the Beckman Young Investigator Award, the Johns Hopkins Alumni Association Excellence in Teaching Award, and the National Science Foundation’s CAREER Award. In 2016 he was elected to the College of Fellows of the American Institute of Medical and Biological Engineering (AIMBE), and he was awarded AIChE’s David Himmelblau Award for Innovations in Computer-Based Chemical Engineering Education. He serves on the editorial board of Proteins, on the scientific advisory board of the Rosetta Design Group, and from 2005-2015 he served on the board of directors of the Ingenuity Project. He is the Diversity Chair of the Rosetta Commons and the Director of the NSF-supported Rosetta Commons Summer Intern Program.
Links: Gray lab site - @jeffreyjgray
What does it take to do impactful science? Solving problems in the 21st century often requires multiple scientists across disciplines to unite under a common goal. These cross-disciplinary collaborations are critical for biomolecular modeling: to tackle problems like understanding the molecular determinants of Alzheimer’s disease and HIV, designing vaccines or drugs or biomolecular materials, and crowdsourcing RNA folding predictions, we require insights from physics, computer science, chemistry, biology, statistics, and more. Yet, most undergraduate research experiences represent a small piece of the puzzle: students work alongside a scientist in a specific field to address a question. To bridge this gap, we created a new NSF-funded Research Experience for Undergraduates (REU) designed to train interdisciplinary, collaborative scientists in biomolecular modeling.
Our research program takes place in the Rosetta Commons, a world-wide consortium of over fifty labs that co-develop the Rosetta software for biomolecular structure prediction and design. United by a common codebase, Rosetta scientists tackle a variety of problems, ranging from solving the structures of proteins and RNA, to design of proteins, interfaces, protein nanomaterials, mineral binders, and antibodies. Our summer program is designed with the goal of immersing students in our community: the students develop new code, perform biomolecular modeling research, and are exposed to the distributed but interconnected research environment where powerful science arises from the synergies of multiple labs.
Selected achievements with Rosetta: (A) antibody structures predicted de novo with sub-angstrom loop conformations, (B) de novo design of an enzyme, (C) proof-of-principle for epitope-focused vaccine design, (D) design of a high-affinity ligand binding pocket, (E) design of self-assembling protein cages, (F) design of peptides that bind and shape inorganic minerals.
Rosetta REU students start their summer at UNC Chapel Hill with a one-week intensive workshop. During this week, students learn principles of software design, dive into the Rosetta libraries, gain exposure to various modeling protocols, and practice programming in Python, XML, shell scripts, and C++. This workshop breaks down the barrier to entry in an interdisciplinary field because students with different expertise can work together to fill in knowledge gaps and bond over learning the core ideas of biomolecular structure prediction and design.
After boot camp, students spend eight weeks in their labs, while remaining connected via weekly virtual journal clubs and cyber-collaboration tools such as Skype, Google Hangouts, and GitHub. These connections show students the benefit of a cyber-linked community to the exchange and development of scientific ideas. Then, students reconvene at the Rosetta Commons summer conference in Leavenworth, WA. Each student presents a poster on their project where they can receive feedback from their cohort and scientists in the Rosetta Community. Here, students also attend plenary sessions, break out discussions, and even group coding sessions that are critical for driving science in the Rosetta community forward.
The REU training has enabled students to participate in highly interdisciplinary projects. One student, Morgan Nance from UC Davis, worked in the Gray lab (Johns Hopkins) to model the structure of sugars on the Fc-FgRIIIa antibody-receptor complex (see figure) important for the binding specificity in proper immune responses. Alexandra Boukhvalova from University of Maryland worked in the Schief lab (Scripps) to design and test modifications to the HIV binding antibody. And Sidney Lisanza of UNC worked in the Baker lab (University of Washington) on self-assembling nanomaterials. Other students in the program worked on projects with greater focus on core development of Rosetta. For example, Ann Cirincione from University of Maryland at Baltimore County worked in the Kuhlman lab (UNC Chapel Hill) on a better algorithm for detecting hydrogen bonds in proteins with applications to interface design, and Jackie Gaston of Carnegie Mellon worked in the Cooper lab (Northeastern U.) on human-computer interface designs to engage FoldIt game players.
So far, we have hosted two cohorts of interns with overwhelmingly positive feedback. Past participants reported that the interdisciplinary Rosetta REU has been formative for their career aspirations. Several of the students noted that the Rosetta REU provided a rare training experience by integrating both computation and wet lab experiments. One student appreciated the community: “The networking opportunity I had is invaluable, I know contacts across the country…the fact that I was treated like a graduate student vastly improved my confidence in research” Of the seven students that have completed undergraduate degrees, four started Ph.D. programs in STEM fields in the Fall of 2015. One student returned as a teaching assistant for boot camp. Another student even returned for a Ph.D., in her summer internship lab.
Another important goal of the Rosetta REU is to increase diversity in the biomolecular modeling community. Women and minorities are underrepresented in science and technology, and especially in fields that emphasize computing. Toward this goal, we recruit students at affinity conferences including the Grace Hopper Celebration of Women in Computing and the Annual Biomedical Research Conference for Minority Students (ABRCMS). At these conferences, several Rosetta Commons scientists and REU alumni helped to recruit for future REU programs.
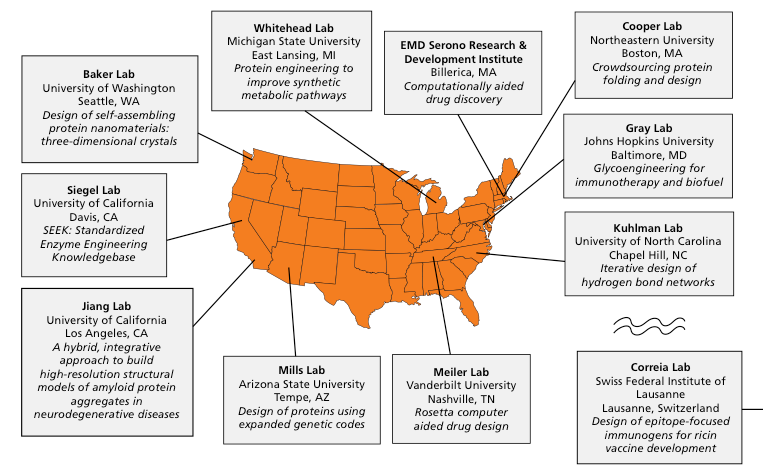
We are looking forward to our next summer program in 2017. This summer, REU students will again work in labs around the world, including the University of Washington, Johns Hopkins, Northeastern, Arizona State, UCLA, UNC Chapel Hill, and the Ecole Polytechnique Federale de Lausanne (EPFL). One student will engage in the pharmaceutical industry at EMD Serono Research & Development Institutein Bilerca, MA. The project topics this year are diverse: ranging from computer-aided drug design to the design of self-assembling nanomaterials. We welcome undergraduates from varied STEM backgrounds, and we are committed to recruiting a diverse cohort. This year’s REU will run from June 5th, 2017 to August 9th, 2017 with applications due on February 1st, 2017. More information about our REU program, including available projects, host labs, and frequently asked questions, can be found at https://www.rosettacommons.org/intern.
 Acknowledgments: The Rosetta summer internship program is funded by an EAGER award from NSF-BIO, grant #1541278.
Acknowledgments: The Rosetta summer internship program is funded by an EAGER award from NSF-BIO, grant #1541278.