RCSB PDB's website at rcsb.org was visited each month by an average of 347,436 unique visitors and 802,076 unique visits. More than 3 TB of data were accessed. Traffic is tracked using AWstats. RCSB.org is visited by >1 million unique visitors/year.
| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2016 | 312,991 | 740,421 | 2381.32 GB |
| February 2016 | 329,488 | 749,712 | 2800.41 GB |
| March 2016 | 359,498 | 845,223 | 3900.47 GB |
| April 2016 | 378,006 | 871,085 | 3679.44 GB |
| May 2016 | 356,342 | 844,134 | 3379.94 GB |
| June 2016 | 326,771 | 802,741 | 3186.85 GB |
| July 2016 | 308,792 | 749,097 | 2399.66 GB |
| August 2016 | 324,314 | 726,959 | 2190.43 GB |
| September 2016 | 354,836 | 789,573 | 2190.57 GB |
| October 2016 | 396,116 | 871,157 | 2457.98 GB |
| November 2016 | 380,632 | 858,851 | 2484.69 GB |
| December 2016 | 341,447 | 775,956 | 1831.00 GB |
A snapshot of the PDB archive (ftp://ftp.wwpdb.org) as of January 1, 2017 has been added to ftp://snapshots.wwpdb.org/. Snapshots have been archived annually since January 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20170101 includes the 125,463 experimentally-determined coordinate files and related experimental data available at that time. Coordinate data are available in PDBx/mmCIF, PDB, and XML file formats. The date and time stamp of each file indicates the last time the file was modified. The snapshot is 757 GB.

The faustovirus (PDB ID 5j7v) is the largest known virus in the PDB
Fast, interactive display of molecular complexes containing millions of atoms is now possible, without plug-ins, on desktop computers and smartphones. The new NGL 3D Viewer enables easy visualization of even the largest structures for everyone in research and education.
In addition, the new binary compressed Macromolecular Transmission Format (MMTF.rcsb.org) massively reduces network transfer and parsing times. With these advances, we are prepared for upcoming advances in experimental techniques that are already delivering structures of rapidly increasing size and complexity.
Sigma-weighted 2m|Fo|-d|Fc| electron density "mini-maps" for ligands are available from the JSmol 3D View. This option is available for ligands with more than one atom (ions excluded) in PDB entries with structure factor data. Different sigma values can be selected.
Watch this brief video for a demo:
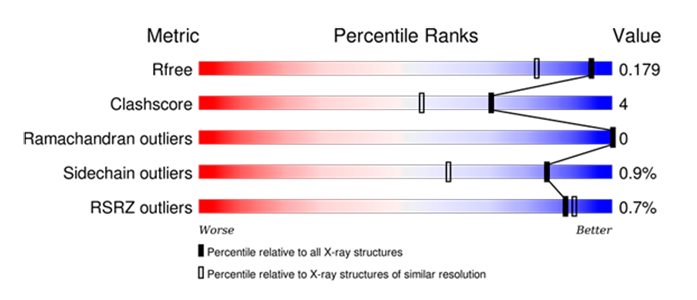
wwPDB Validation Reports are available for every entry to provide an assessment of the quality of a structure and highlight specific concerns by considering the model coordinates, experimental data, and fit between the two.
RCSB PDB Structure Summary pages contain the “slider” graphic from these reports that provides a visual summary and also link to the full PDF report.
New options in the NGL 3D viewer can be used to map wwPDB Validation Report information onto the 3D structure. Clashes can be displayed as pink disks, and the full structure can be shown using “Geometry Quality” and “Density Fit” coloring schemes.
 |
 |
wwPDB Validation Slider graphic for PDB 3WY6. |
Color By Density Fit: Hydrolases colored using the “Density Fit” scheme. |
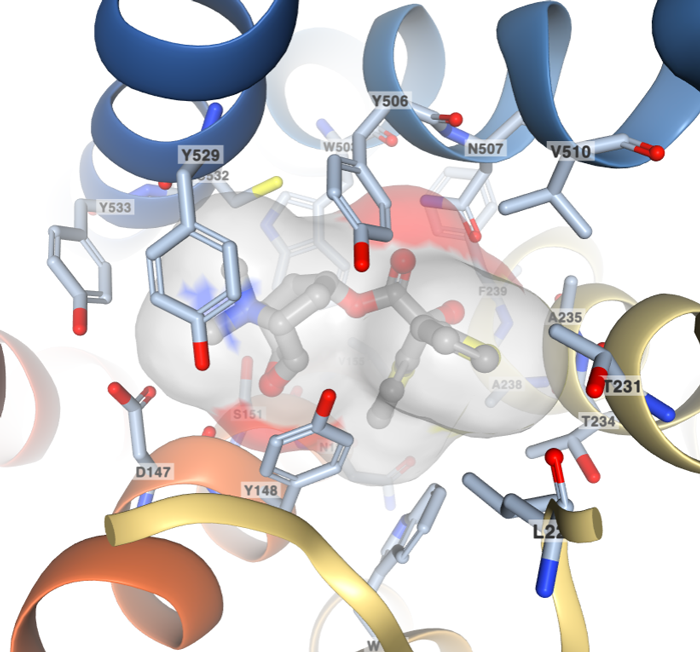
Ligand interaction view of the COPD drug tiotropium (Ligand ID 0HK) bound to the Muscarinic acetylcholine receptor M3 (PDB ID 4U15).
The Small Molecules section of an entry’s Structure Summary page offers multiple tools for ligand visualization in 2D and 3D. A new option uses the 3D NGL viewer to display the ligand molecule and all residues within 5 Ångstrom. The vdW surface of the ligand highlights the occupied volume. The name and number of interacting residues are labeled. From this 3D view, users can access additional ligands in an entry using the Interaction dropdown menu.
RCSB PDB provides additional annotations and query options for transmembrane proteins.
New high resolution images for transmembrane proteins were created using information from the Orientations of Proteins in Membranes (OPM) database.
Molecular graphics and analyses performed with UCSF ChimeraX, developed by the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIGMS P41-GM103311).
Data about the orientation of transmembrane proteins are provided by the Orientations of Proteins in Membranes (OPM) database, when available. OPM is updated less frequently than the PDB, so not all transmembrane proteins will have these images.
 |
 |
 |
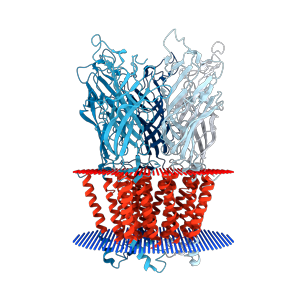 |
Transmembrane views for the pentameric ligand gated ion channel GLIC (PDB ID: 2xq7)
BIRD Summary pages, accessible from the Small Molecules widget on an entry's Structure Summary Page, offer images, high level details, and links to reference files for the small molecules described in the wwPDB Chemical Component Dictionary.
These pages now offer summaries for the peptide-like antibiotic and inhibitor molecules described in the wwPDB Biologically Interesting molecule Reference Dictionary (BIRD), including BIRD class, compound details, and description.
Examples of structures contained in BIRD include actinomycin D and vanomycin.
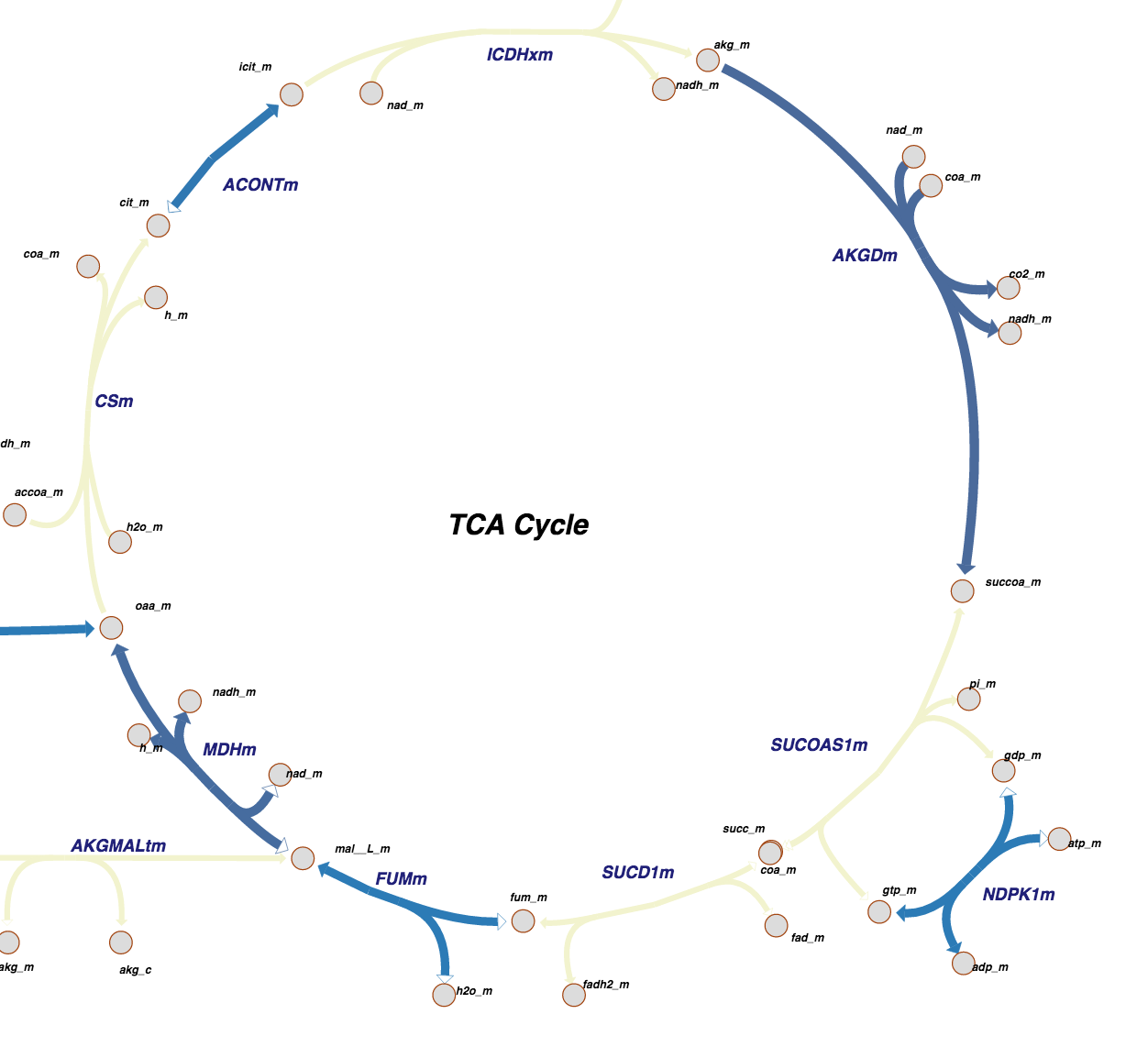
Citric Acid Cycle as seen in Pathway View. The color-coding indicates what parts of the pathway have been determined at atomic level resolution in PDB, inferred via homology models, or cannot be modeled right now.
3D structures of an increasing number of proteins that perform reactions within metabolic pathways are available in the PDB. To accommodate a deeper structural view of biology, Pathway View allows users to browse through well-characterized metabolic pathways and access the corresponding PDB entries.
Access Pathway View from the top menu Visualize options. It has also been integrated into the Structure Summary pages of PDB entries linked to pathways. In addition, the Protein Feature View page displays a map of the relevant pathway portion if the corresponding UniProt sequence is linked to a metabolic pathway.
Protein Modifications in the PDB have been mapped to the RESID database of Protein Modifications and the associated Protein Modifications Ontology (PSI-MOD). Explore these modifications with the new Protein Modification Browser, Advanced Search, and the 3D View and Sequence options on Structure Summary pages. A detailed overview is available.
A new Advanced Search option for Wild Type Protein selects protein sequences that do not contain mutations in comparison with the reference UniProt sequence.
Some PDB entries include expression tags that were was added during the experiment. Select "No" for the "Include Expression Tags" option to filter out sequences with expression tags.
PDB entries may also contain only a portion of the referenced UniProt sequence. The "Percent coverage of UniProt sequence" option defines how much of a UniProt sequence needs to be contained in the PDB entry.
RCSB PDB has been developing the RCSB.org website with mobile users in mind.
The home page, search results, and other pages were designed to be responsive to the devices that display them. For example, smaller tablets and mobile phones display Structure Summary as a menu option rather than tabs that appear across desktop monitors.
The new NGL viewer can render complex molecular machines in 3D, without any plug-ins, on desktops and mobile devices. NGL also uses the new binary compressed Macromolecular Transmission Format (MMTF.rcsb.org), which massively reduces network transfer and parsing times.
With the combination of these advances, the RCSB.org experiences is no longer device-specific. The combination of the NGL viewer and mobile-responsive web design at rcsb.org offers fast and comprehensive on-the-go access on mobile devices. As a result, the mobile app will be retired at the end of 2016.
Questions about these services may be sent to info@rcsb.org.