| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2017 | 351,773 | 763,080 | 1867.92 GB |
| February 2017 | 360,900 | 772,850 | 2454.42 GB |
| March 2017 | 418,335 | 906,522 | 2906.16 GB |
We are looking for talented and highly motivated Postdoctoral Fellows to join our multidisciplinary development team.
The Challenge: Develop innovative analysis, integration, query, and visualization tools for 3D biomolecular structures to help accelerate research and training in biology, medicine, and related disciplines. In these projects, we employ the latest advances in computer science to develop highly interactive features and scalable services and workflows. The overall goal is always to develop highly interactive features for the RCSB PDB website.
Two positions are available: one focused on Visualization (position #1104) and Machine Learning/Biological Assemblies (position #1105).
Possible project areas include:
This is a unique opportunity to engage in leading edge research, development, and outreach activities of the RCSB PDB with worldwide impact. For more, visit out Careers page or Contact Us with questions.
3D structures of an increasing number of proteins that perform reactions within metabolic pathways are available in the PDB. To accommodate a deeper structural view of biology, Pathway View allows users to browse through well-characterized metabolic pathways and access the corresponding PDB entries.
Access Pathway View from the top menu Visualize options. It has also been integrated into the Structure Summary pages of PDB entries linked to pathways. In addition, the Protein Feature View page displays a map of the relevant pathway portion if the corresponding UniProt sequence is linked to a metabolic pathway. Additional information is available.
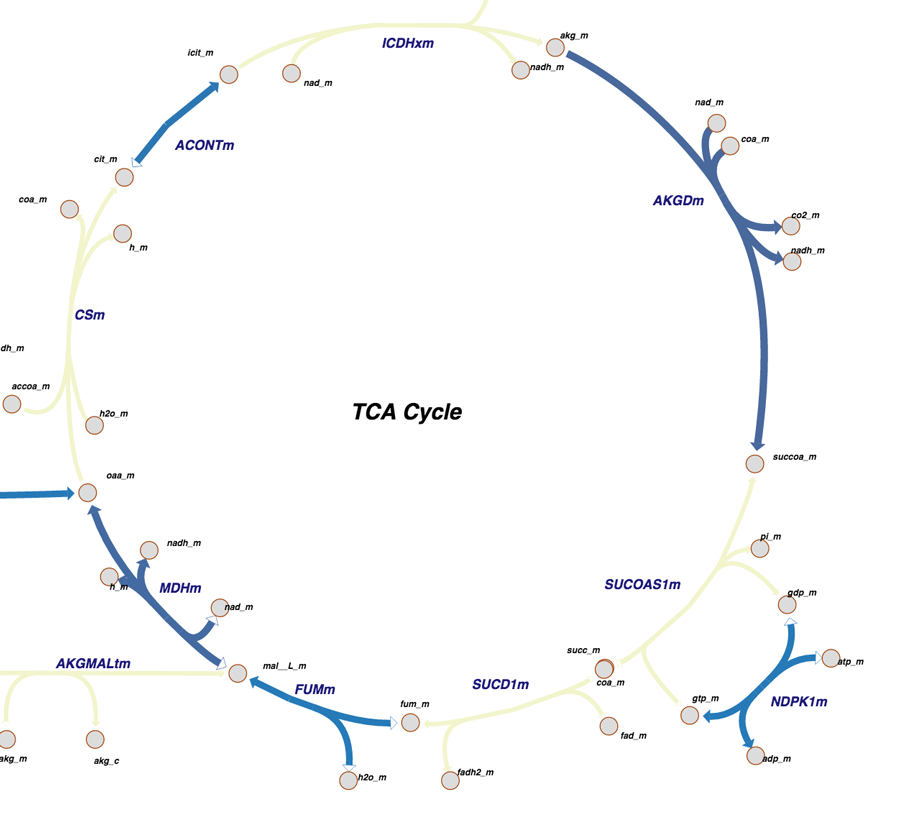
Citric Acid Cycle as seen in Pathway View. The color-coding indicates what parts of the pathway have been determined at atomic level resolution in PDB, inferred via homology models, or cannot be modeled right now.
The Citric Acid Cycle was also highlighted by the Molecule of the Month.
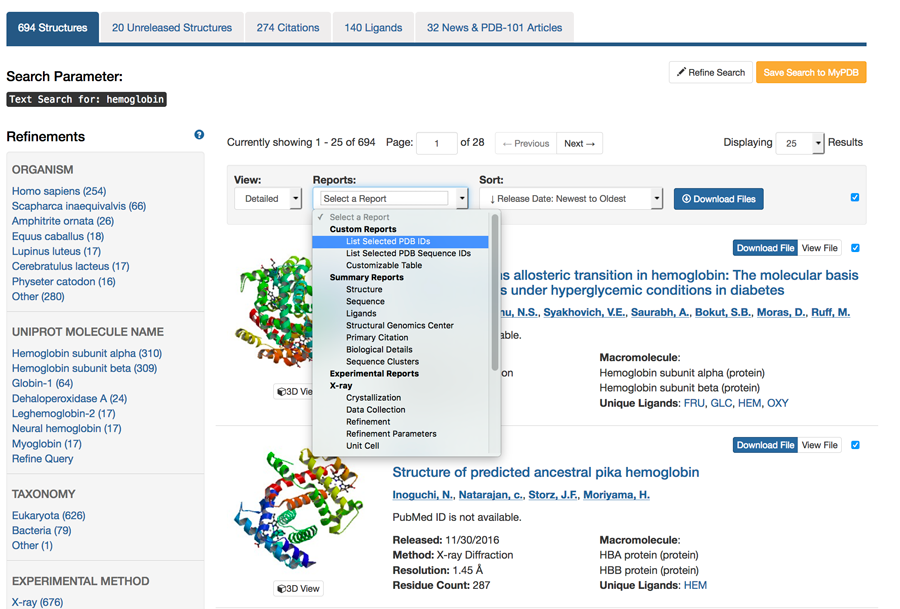
Did you ever wish you could get just a list of PDB IDs from your search? You can! From the results page of any search that returns multiple entries, the ‘Reports’ menu offers an option to ‘List Selected IDs’ to get a list of the PDB IDs from your results set. The list will appear in a new tab in your browser.
Other Report options include creating tabular reports that can then be filtered by content, sorted, and downloaded as an Excel spreadsheet.
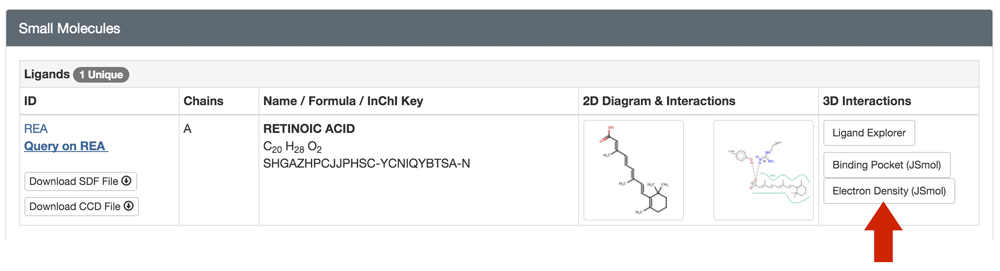
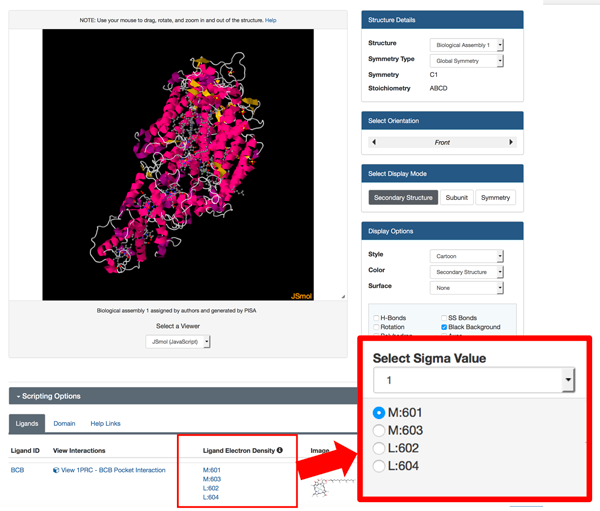
View sigma-weighted 2m|Fo|-d|Fc| electron density "mini-maps" for bound ligands using the JSmol 3D View. This feature is available for ligands with more than one atom (ions excluded) in PDB entries with structure factor data. Different sigma values can be selected.
The feature can be accessed from an entry's Structure Summary page in the Small Molecule's section or from the bottom of the 3D View page.
 |
 |
| Electron density mini-map for Thymidine-5'-Phosphate in PDB ID 3HW4 at sigma level 2. Electron density is only available for part of the ligand. | Retinoic Acid in PDB ID 1CBS at sigma level 1. In this example, the ligand fits well into the density. |
 |
|
| Electron Density (JSmol) button from the Structure Summary page for PDB entry 1CBS. |
 |
|
| Options to view different ligands and mini-maps and to change sigma levels appear under the JSmol viewer for PDB entry 1PRC. |

The article The RCSB Protein Data Bank: integrative view of protein, gene and 3D structural information describes the new tools and resources have been added to the RCSB PDB web portal in support of a ‘Structural View of Biology.’ Recent developments have improved the User experience, including the high-speed NGL Viewer that provides 3D molecular visualization in any web browser, improved support for data file download and enhanced organization of website pages for query, reporting and individual structure exploration. Structure validation information is now visible for all archival entries. PDB data have been integrated with external biological resources, including chromosomal position within the human genome; protein modifications; and metabolic pathways. PDB-101 educational materials have been reorganized into a searchable website and expanded to include new features such as the Geis Digital Archive.
The RCSB Protein Data Bank: integrative view of protein, gene and 3D structural information
Peter W. Rose, Andreas Prlic, Ali Altunkaya, Chunxiao Bi, Anthony R. Bradley, Cole H. Christie, Luigi Di Costanzo, Jose M. Duarte, Shuchismita Dutta, Zukang Feng, Rachel Kramer Green, David S. Goodsell, Brian Hudson, Tara Kalro, Robert Lowe, Ezra Peisach, Christopher Randle, Alexander S. Rose, Chenghua Shao, Yi-Ping Tao, Yana Valasatava, Maria Voigt, John D. Westbrook, Jesse Woo, Huangwang Yang, Jasmine Y. Young, Christine Zardecki, Helen M. Berman, and Stephen K. Burley
Nucleic Acids Research (2017) 45 (D1): D271-D281 doi:10.1093/nar/gkw1000
A list of all RCSB PDB publications and information on citing RCSB PDB and related resources is available.