Data Exploration Services
RCSB.org Statistics
Website traffic is tracked using AWstats. RCSB.org is visited by >1 million unique visitors/year.
| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2018 | 469,866 | 1,308,127 | 3123.22 GB |
| February 2018 | 434,550 | 1,208,166 | 2719.71 GB |
| March 2018 | 500,382 | 1,411,474 | 2578.50 GB |
New Architecture and Services Enable Faster Access to More Information
Recent architecture improvements provide faster access to RCSB.org content with improved page load times. Some of the more prominent changes are summarized below.
Simplified URL Structure
RCSB PDB is moving to a new microservice-based architecture to better scale our services to accommodate growth and user demand, and to increase the speed at which we can deploy new services. In parallel, we are streamlining our URLs for easy access and sharing. The previous URL structure is still fully supported (for the time being) and will automatically redirect to the new URLs. Examples are below
Old URL Structure
- Home Page https://www.rcsb.org/pdb/home/home.do
- SSP https://www.rcsb.org/pdb/explore/explore.do?structureId=4dkl
- 3D View http://www.rcsb.org/pdb/ngl/ngl.do?pdbid=4dkl&bionumber=1
- Careers https://www.rcsb.org/pdb/static.do?p=general_information/about_pdb/contact/job_listings.html
New URL Structure
- Home Page https://www.rcsb.org/
- SSP https://www.rcsb.org/structure/4dkl
- 3D View https://www.rcsb.org/3d-view/4dkl/1
- Careers https://www.rcsb.org/pages/jobs
Changes to the Structure Summary Page (SSP)
Top-level SSP pages utilize the new microservice architecture to enable fast access to:
- Structure funding information
- Macromolecule sequence information
- Comprehensive symmetry information
- Biological assembly evidence for recent structures
- Software packages used
- Deposition identifiers for large groups of related structures submitted
PDB Statistics
Improved display of PDB statistics are available; including the distribution of structures released per year, and refinement software used.
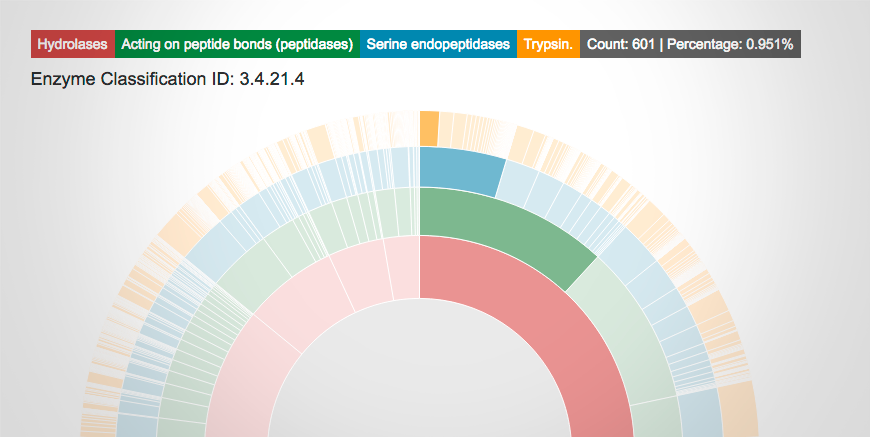
The new visualization of PDB Data Distribution by Enzyme Classification allows users to interactively explore the enzyme classes in the PDB.
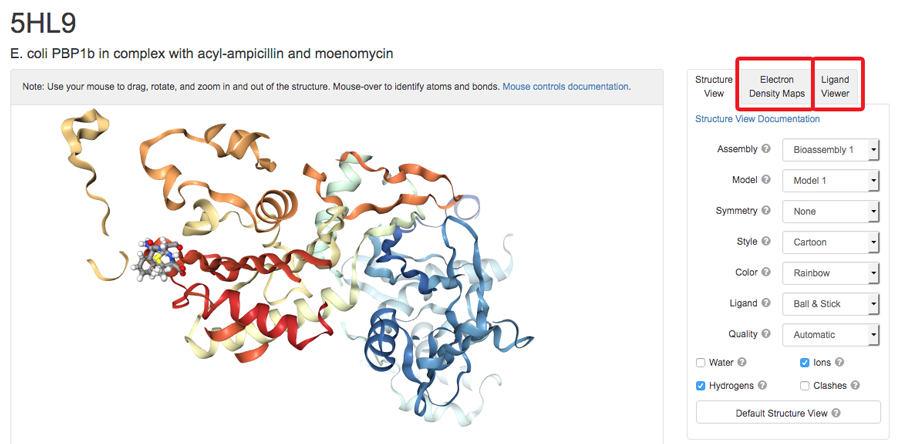
View Interactions and Maps in 3D
The 3D View now offers options for macromolecular-ligand interactions and X-ray electron density maps using NGL. Ligand interaction viewing options include hydrogen bonds, hydrophobic contacts, halogen bonds, pi interactions, and metal interactions. A 3D View User Guide is available.
3D View: Ligand Viewer
Features include options to display the binding pocket surface and non-covalent interactions (Hydrophobic Contacts, Hydrogen Bonds, Halogen Bonds, Metal Interactions, Pi Interactions).
Calculations are performed in real-time within the web browser using the NGL Viewer, our web-based tool for molecular graphics and structure analysis.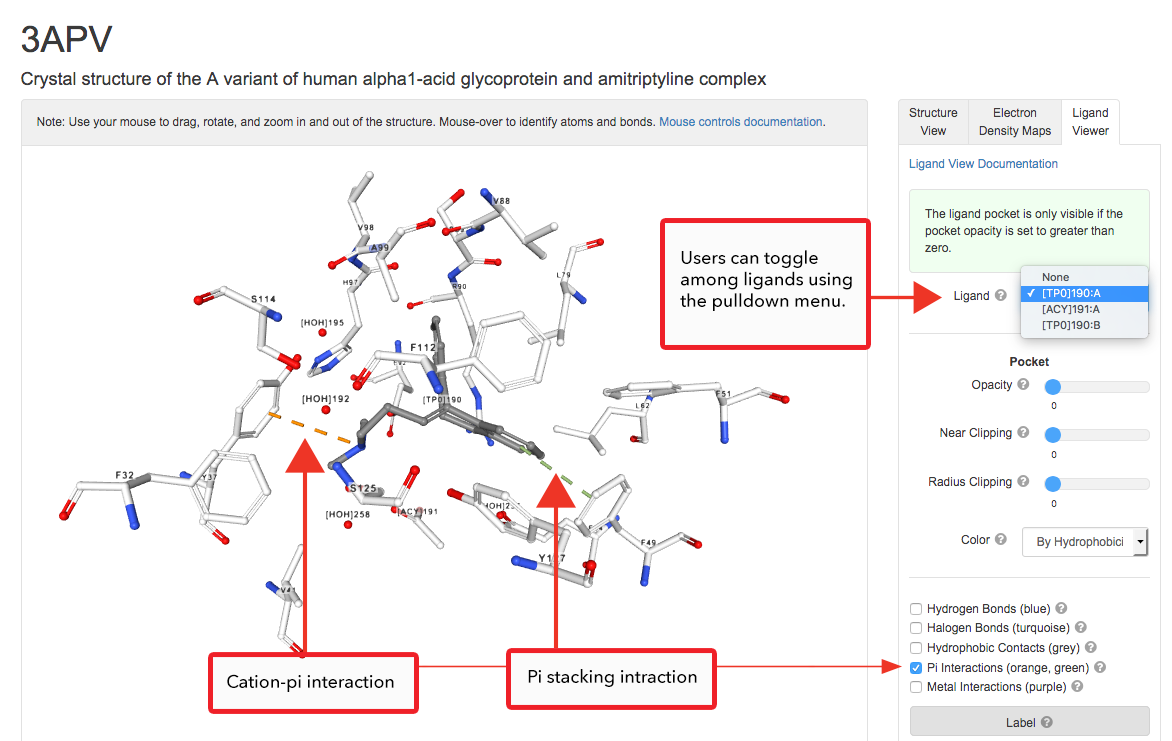
Pi stacking and Cation-pi interaction of amitriptyline (ligand ID TP0) as seen in the A variant of human alfa1-acid glycoprotein (PDB structure 3APV).
3D View: Electron Density Maps
NGL now displays 2fo-fc (blue mesh/surface) and fo-fc (red/green mesh/surface) maps in the context of the structural model for X-ray crystallographic structures. Electron density maps combine the structural model (coordinates) and the experimentally-collected data from an X-ray structure determination and serve to represent the fit of the model to the data. Even in the best quality structures, there are areas of poor electron density, which may represent sections of the model that exist in multiple conformations. For more information, see the User Guide or learn more about Structure Factors and Electron Density at PDB-101.
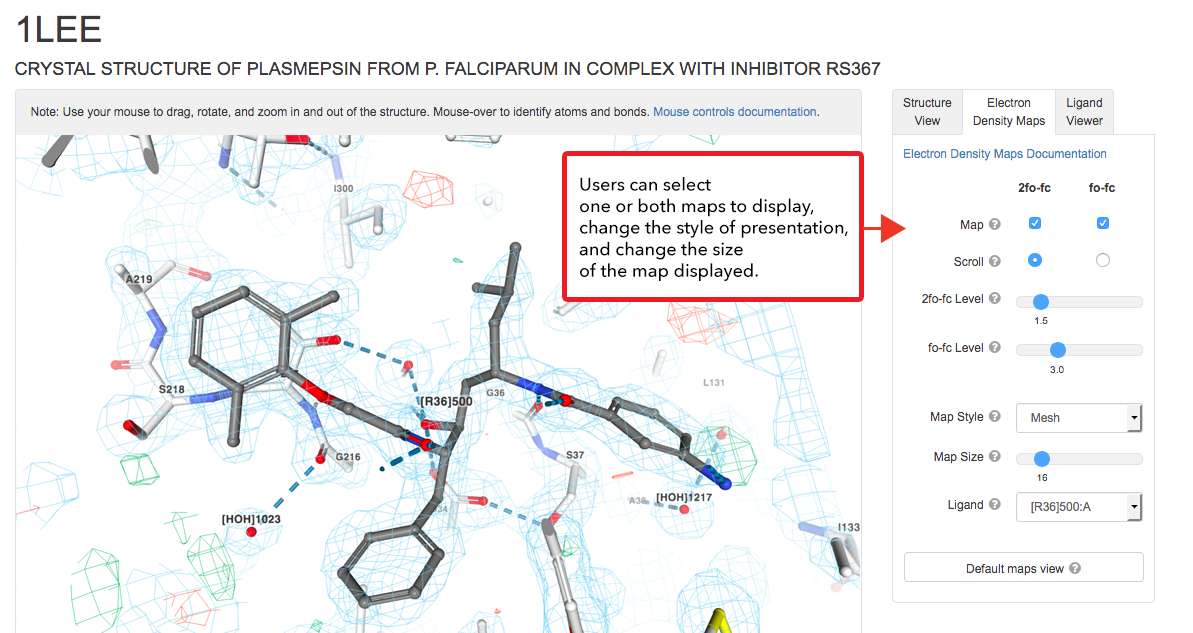
Close up view of the electron density around the inhibitor (ligand ID R36) seen in complex with plasmepsin (PDB structure 1LEE; zoom in to get same view).
Postdocs and Developers: Join Our Team
RCSB PDB is looking for Front End Developers (Rutgers) and Postdoctoral Fellows (UCSD) to join our multidisciplinary development team.
The Challenge: Develop innovative analysis, integration, query, and visualization tools for 3D biomolecular structures to help accelerate research and training in biology, medicine, and related disciplines. In these projects, we employ the latest advances in computer science to develop highly interactive features and scalable services and workflows. This is a unique opportunity to engage in leading edge research, development, and outreach activities of the RCSB PDB with worldwide impact. For more, visit our Careers page or Contact Us with questions.
