Outreach and Education 
Browse Molecular Evolution at PDB-101
PDB-101 is an online portal for teachers, students, and the general public to promote exploration in the world of proteins and nucleic acids. PDB-101 features support learning about the diverse shapes and functions of these biological macromolecules and their relationship to biomedicine and agriculture, from protein synthesis to health and disease to biological energy.
Frances Arnold, a winner of the 2018 Nobel Prize in Chemistry, pioneered the use of evolution to create enzymes with entirely new functions. This has inspired a new Molecule of the Month article on the Directed Evolution of Enzymes and a new Browse section that collects all PDB-101 materials related to Molecular Evolution.
The diverse plants, animals, and other living things that currently populate the Earth are thought to have evolved from a common primordial ancestor. Atomic structures have revealed the molecular mechanisms of evolution, showing how molecules change by random mutation and recombination, and are selected for optimal function. Learn more about these structures at PDB-101.

|

|
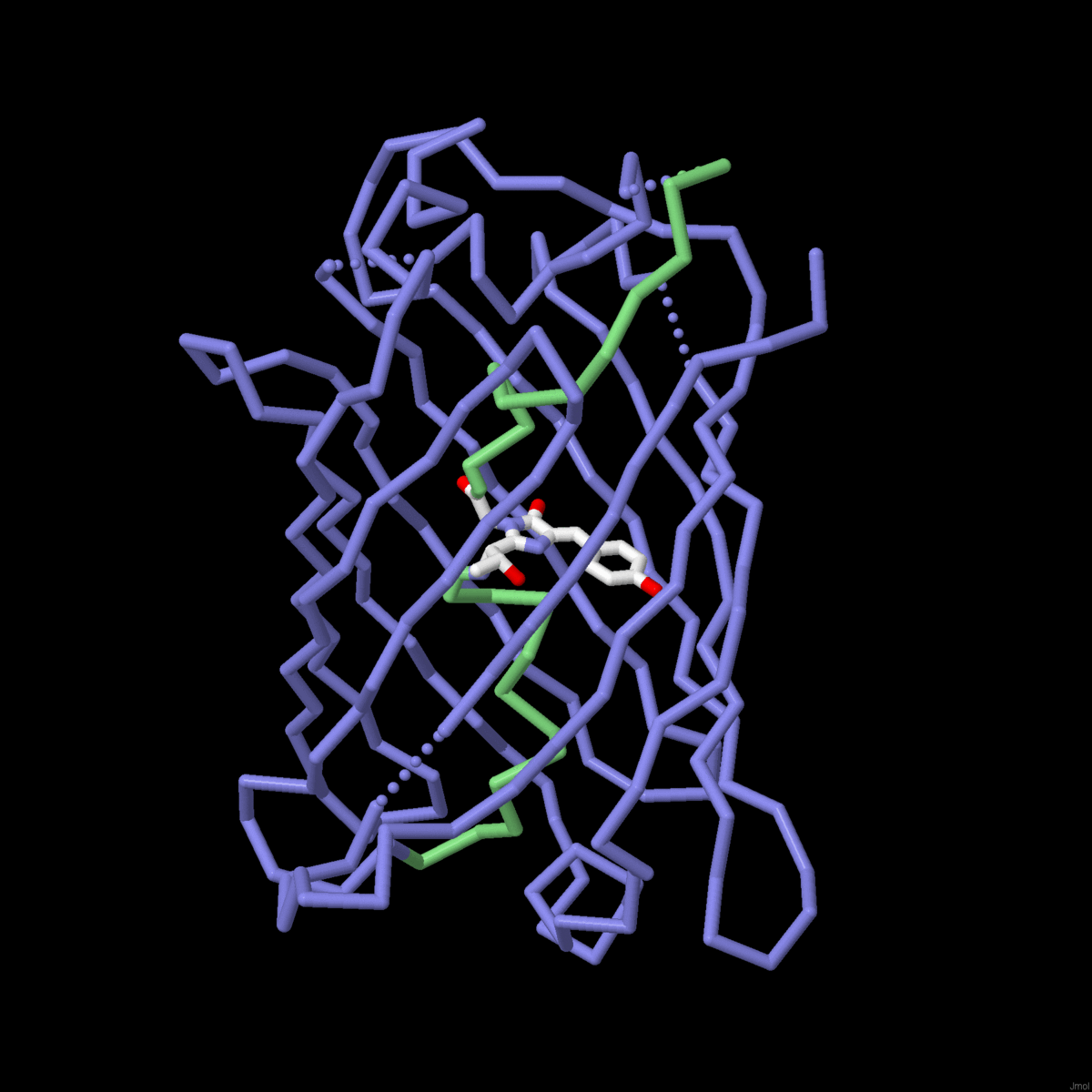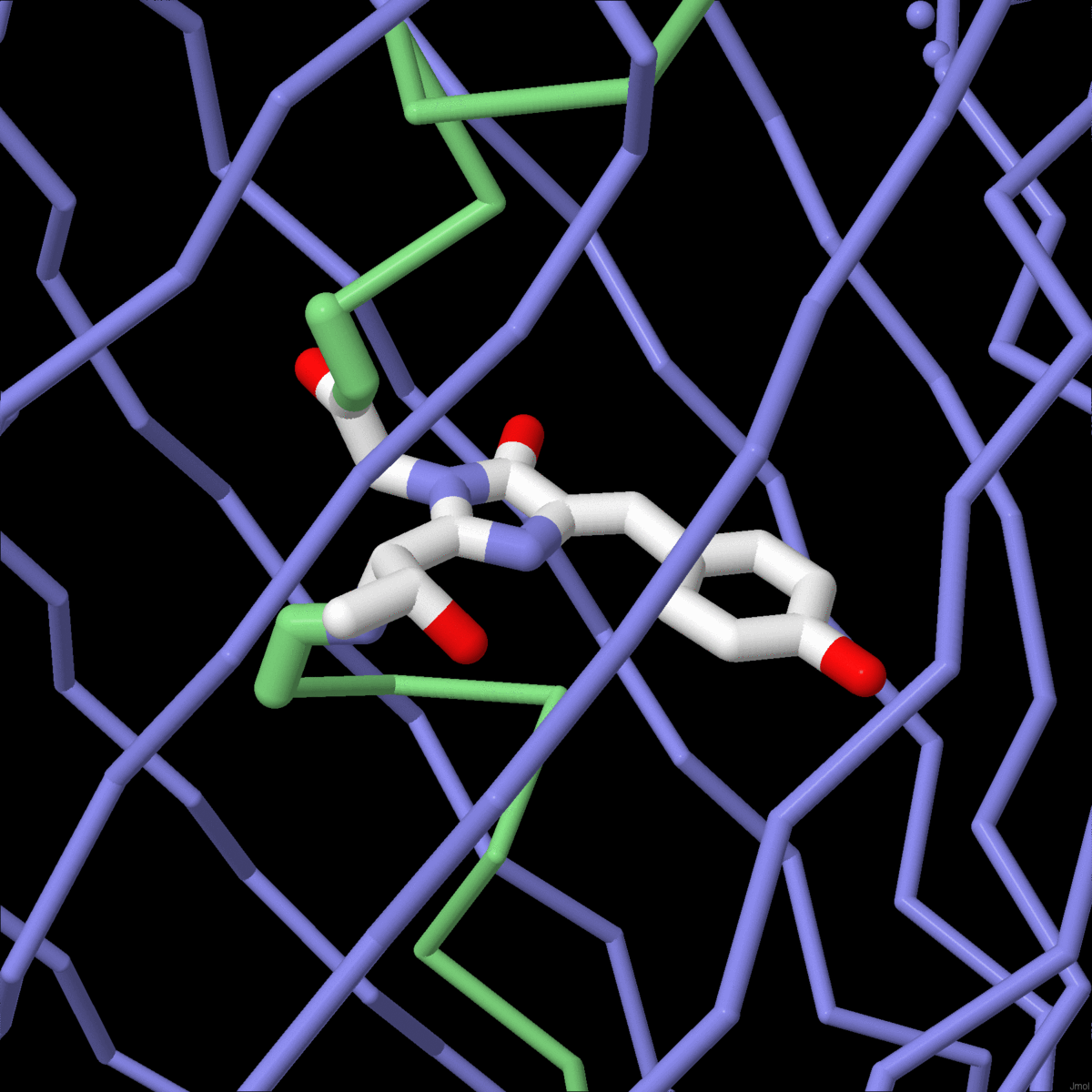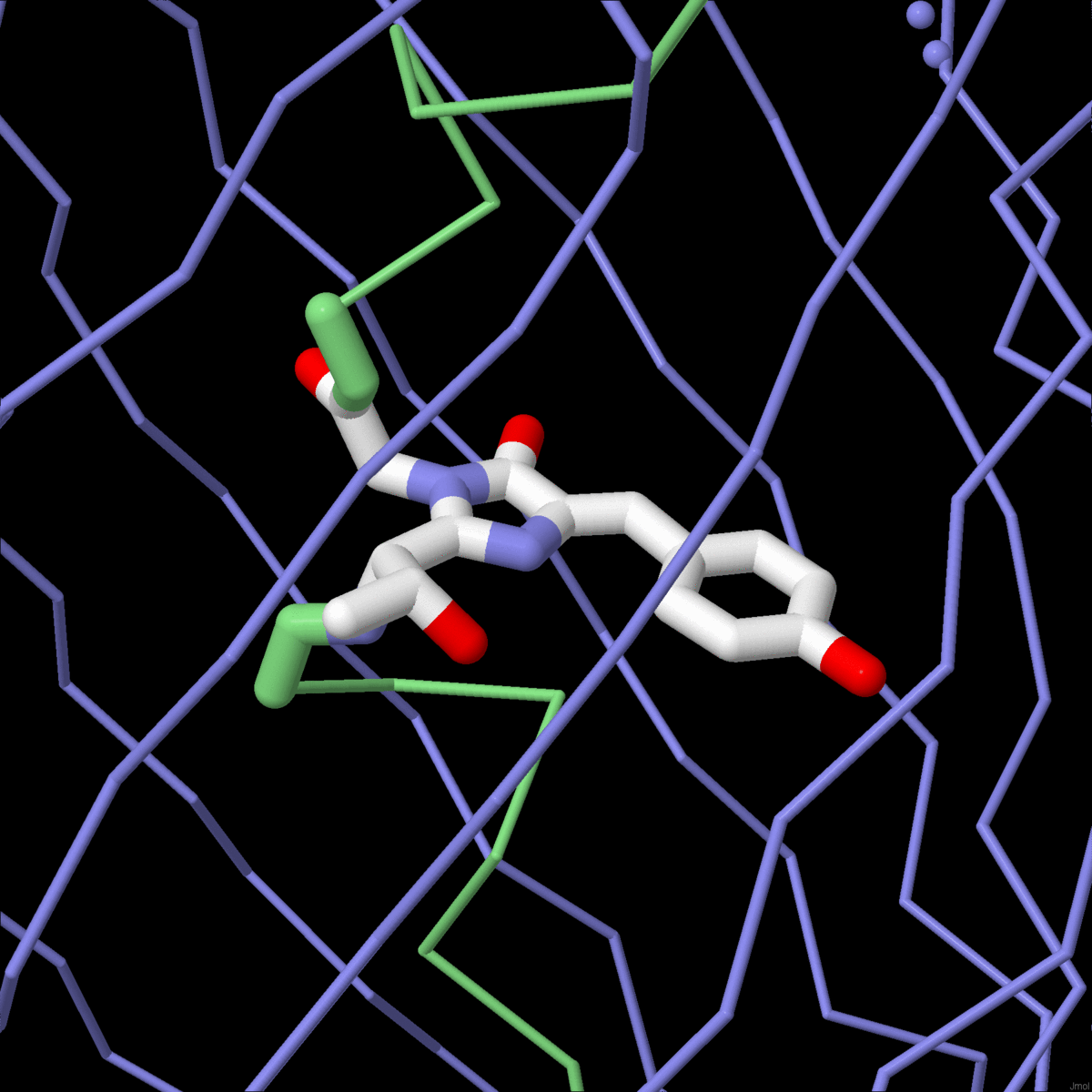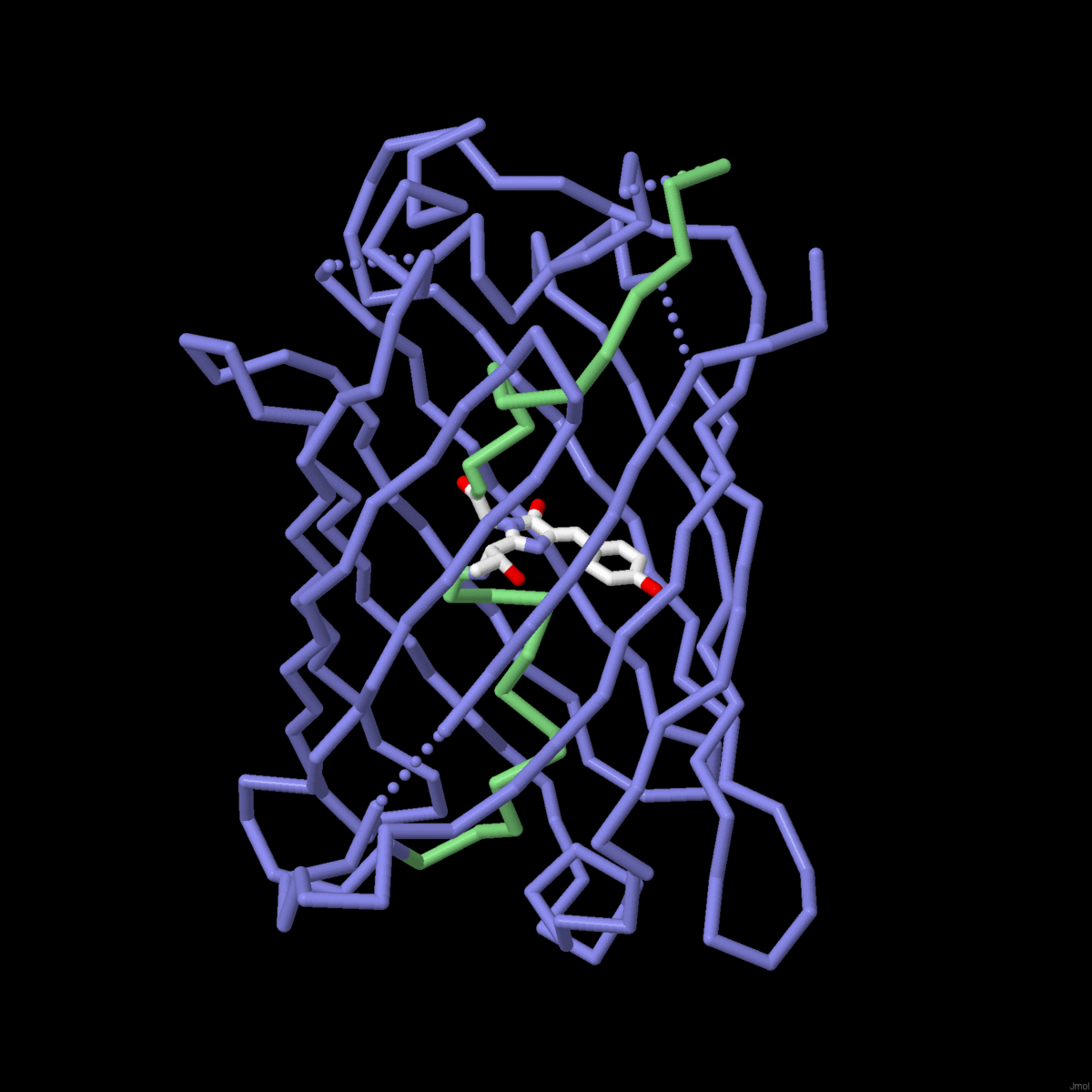
| The mechanisms of molecular evolution are revealed in globin sequences and structures. |
Molecule of the Month: Biological evolution is being harnessed in the lab to create new enzymes. |
Contact Customer Services with Questions and Feedback
RCSB PDB's Customer Services collects and answers questions about the website, PDB data, and structural biology. Nearly 1,000 unique users initiate new electronic conversations each year.
Questions and comments come from students new to structural biology, users involved in the general study of science, and domain experts from the various disciplines that utilize PDB data.
Related resources include Help Topics for a variety of RCSB PDB services, Website FAQ, Guide to Understanding PDB Data, and a Service Status page that indicates if all online services are up and running.
Superbugs and Antimicrobial Resistance
Since the discovery of penicillin, researchers have developed better and better antibiotics to fight microbial infection. Bacteria, in turn, have evolved more and more effective ways of resisting these drugs: by destroying them, by pumping them out, or by modifying the target of the drug. Atomic structures help us understand resistance to antibiotics and develop new ways to fight infection.
PDB-101 now hosts new videos and a poster that explore antimicrobial resistance (AMR) at the molecular level. The poster Superbugs! How Bacteria Evolve Resistance to Antibiotics highlights the protein structures that medical researchers are using to search for ways to fight these superbugs while new videos explore Staphylococcus aureus and its resistance mechanisms to penicillin and other beta-lactam antibiotics.
The 2018 Video Challenge inspired instructive videos created by high school students that presented the Mechanisms of Bacterial Resistance to Beta-lactam Antibiotics.
Mechanisms of Bacterial Resistance to Aminoglycoside Antibiotics is focus of the 2019 Video Challenge. High schools students are asked to tell a story that communicates the molecular changes that occur in bacteria that help them to become resistant to aminoglycoside antibiotics using relevant 3D protein structures. Videos should also address the dangerously high level of antibiotic resistance caused by misuse and overuse of antibiotics. Entries can be submitted as early as January 15, 2019.
The AMR Browse category links to all AMR-related resources at PDB-101.
Curated 3D Views at PDB-101
Written and illustrated by David S. Goodsell, Molecule of the Month features provide an easy introduction to PDB structures for teachers and students around the world.
Since January 2000, this series has explored the structure and function of biomacromolecules from AAA+ Proteases to Zika Virus. Each installment includes an introduction to the structure and function of the molecule, a discussion of the relevance of the molecule to human health and welfare. Most articles include interactive 3D views in JSmol to highlight specific structural features.
New 3D views have been added to popular articles including Collagen, Lysozyme, DNA, Ferritin, Hemoglobin, and Catalase. These features were handcrafted by RCSB PDB intern Belle Lin. Explore one of these new features today!
Create a Nobel-Themed Bean Bag Toss
In 1962, Max Ferdinand Perutz and John Cowdery Kendrew received the Nobel Prize in Chemistry "for their studies of the structures of globular proteins."
HemoHole is a game where hemoglobin has holes where the heme bean-bags can be tossed through. It was created at Rutgers by Institute for Quantitative Biomedicine graduate students Kristin Blacklock, Elliott Dolan, Will Hansen, Nancy Hernandez, and Dmitri Zorine.
Visit PDB-101 to download the SVG files used to create HemoHole and to learn more about hemoglobin, myoglobin, PDB Pioneers, and Structural Biology and Nobel Prizes.
 |
 |
| The hemoglobin image was broken into two SVG files and laser cut/etched. |
HemoHole is a game where hemoglobin has holes where the heme bean-bags can be tossed through. |
2018 Papers Published
RCSB Protein Data Bank: Sustaining a living digital data resource that enables breakthroughs in scientific research and biomedical education (2018) Protein Science 27: 316–330 doi: 10.1002/pro.3331
From atoms to cells: Using mesoscale landscapes to construct visual narratives (2018) Journal of Molecular Biology 430: 3954–3968 doi: 10.1016/j.jmb.2018.06.009
Molecular illustration in research and education: Past, present, and future (2018) Journal of Molecular Biology 430: 3969-3981 doi: 10.1016/j.jmb.2018.04.043
Learning biology through molecular storytelling (2018) The Science Teacher 86: 28-33
Analysis of impact metrics for the Protein Data Bank (2018) Scientific Data 5: 180212 doi: 10.1038/sdata.2018.212
Amino acid modifications for conformationally constraining naturally occurring and engineered peptide backbones: Insights from the Protein Data Bank (2018) Biopolymers 109: e23230 doi: 10.1002/bip.23230