Education Corner
Exploring the Molecules of Biological Warfare in Virtual Reality
By Steve McCloskey, Nanome Inc.
Steve McCloskey is an alumni from the first class of Nanoengineering at the University of California, San Diego. Steve’s work is focused on emerging technologies applied to Science, Technology, Engineering, and Mathematics (STEM).
During his time at UC San Diego, Steve worked directly with the founding Chair of the Nanoengineering Department, Ken Vecchio helping set the foundation for the Nanoengineering Materials Research Center and developing thermodynamic processing methods for Iron-based Superelastic alloys.
After graduating from UCSD he founded Nanome Inc to build Virtual Reality solutions for Scientists and Engineers working at the nanoscale, specifically protein engineering and small molecule drug development.
About Nanome
San Diego-based Nanome Inc., which created the first molecular modeling app for modern Virtual Reality (VR) systems, has released its flagship software (Nanome) for free download on the Oculus, Steam, and Viveport stores. Nanome opens the door to a virtual world where users can experiment, design, and learn at the nanoscale alongside colleagues and friends.
Nanome imports molecular structures directly from resources such as RCSB PDB, PubChem, and DrugBank. Users can measure distances and angles between atoms, mutate amino acids, design new molecules, and much more. Nanome promotes seamless collaboration on research, classroom presentations, or homework. Critical research findings can be presented in a virtual world alongside colleagues, friends, and audiences around the world.
Nanome has worked with RCSB PDB to offer an ever-changing "Featured Molecules" menu that highlights structures featured in the Molecule of the Month series along with selected small molecules.
Here at Nanome, we believe the most important educational opportunities come from contributing to medical cures or preventing biological warfare. That’s why we decided to share a story about discovery and education that could improve our national security.
On March 4, 2018, Sergei Skripal and his daughter Yulia Skripal were poisoned by a fine powdery substance in Salisbury, England. The British government accused Russia of attempted murder on March 12 and expelled at least 23 diplomats two days later. Both Sergei and Yulia survived the poisoning.
Sergei is a former Russian military officer and double agent for the UK’s intelligence services. UK sources and the Organisation for the Prohibition of Chemical Weapons (OPCW) identified a Novichok nerve agent (A-234) as the poison. Not much is known about how A-234 might have afflicted Sergei and Yulia at the molecular level in a way to be resistant to most commonly used antidotes.
UC San Diego professor and Nanome advisor Zoran Radić, an expert on such compounds, developed a hypothesis about how A-234 poisoned Sergei and his daughter at the molecular level. He used Nanome’s VR molecular visualization software to explore Novichok A-234 in 3D.
Radić has been studying acetylcholinesterase (AChE) structure and function for the past 40 years. "This key enzyme in our metabolism is the known target of organophosphate toxicants and Novichoks belong to that category of compounds," he told Nanome. Currently, as a PI, he leads a major multi-institutional effort funded by the NIH CounterAct Program to create advanced antidotes against organophosphates poisoning caused by insecticides, medications, and nerve agents.
Investigating Novichok in VR
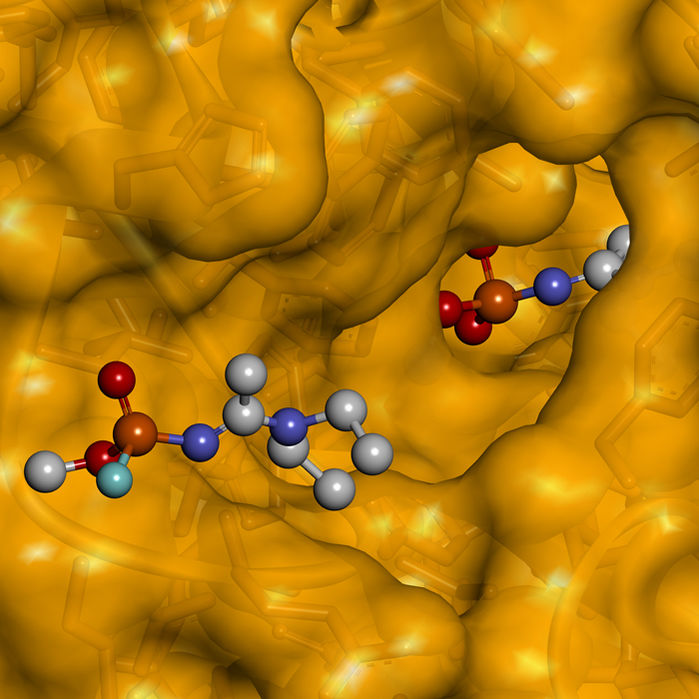
New modeling studies of the nerve agent A-232 (ball and stick) depict its descent from acetylcholinesterase’s gorge opening (left) to a tight fit in the enzyme’s deep active center. Z. Radic, using NanoPro VR by Nanome, Inc.
Since Novichok nerve agents (Novichok means "newcomer" in Russian) originated during the 1970s in the Soviet Union, and only top-security government labs with proper clearance could experiment with the agents, knowledge remains limited to the outside world. Most world governments have been either tight-lipped or have denied possession of and experimenting with Novichoks.
So, Radić turned to information published by Soviet dissidents who had defected to the West and shared Soviet classified knowledge. One particular book described actual structures of some of the Novichok compounds. Publicly available documents supported such sources.
From paper-drawn 2D sketches, he created a coordinate file of A-234 in PDB (Protein Data Bank) format and imported it into Nanome—VR software used by scientists and researchers to visualize complex 3D data by loading in molecules from the RCSB PDB.
"There are a number of very different structures described as Novichoks out there, and understanding the geometry of the enzyme, I was able to pick those that are most likely to be the culprit [behind the poisoning]," said Radić.
Acetylcholinesterase stops the signal between a nerve cell and a muscle cell. Radić hypothesized that if a Novichok bind to acetylcholinesterase, blocking access to known antidotes, is severe damage and possible death could result. He positioned his model of A-234 to the enzyme’s active center and used Nanome ’s minimization feature to adjust the geometry.
"It is relatively simple," he said. "I created models in a classical model building software, saved it in PDB format, then opened it in Nanome."
Nanome optimized the experience, making it easier and more efficient. "When I can stand [right up to] the molecule, complementarity in geometries and electronic features [of the enzyme and the nerve agent] becomes much more obvious. I understand what would fit where. It’s like a 3D Tetris game. This works much faster for me than 2D, 3D-glasses, and other classical approaches to visualization."
He adds: "I can see how this accelerated deduction could easily lead to seeing certain aspects that would otherwise be omitted in a system that doesn't give return information as quickly or deeply and rich in its representation. And the ability to computationally optimize ligand geometry and its interaction with macromolecule in VR in real time comes as an immense bonus."
Presenting Novichok in VR
Radić uses Nanome’s VR tool to present his findings at conferences and meetings, where three or four people can directly interact with molecules in VR workspaces, and large audiences can observe VR interactions via 2D projection on a big screen. "If they want to look at a Novichok structure in VR, they could simply enter into one of the VR-enabled rooms and explore." In the past two years, he has used Nanome for VR presentations at ten international conferences, three university courses, and in daily research.
When presenting how Novichok works to an audience in VR, Dr. Radić first shows his audience the structure of the small compound and the way it is accessing the bound state. "I can do that manually [with Nanome]," he points out, motioning with his hands. "I can take [the molecule] in my hand and put it here or put it there. I usually have a blank rendering of already bound molecules, for a reference. I just go and [select or unselect] and allow the audience to view my guess and the actual bound state."
He believes Nanome represents an instant way of communication. "A Chinese proverb says that an image is worth a thousand words," he said. "VR representation adds another dimension and an order of magnitude to it. Essentially, inherently compressed information is instantaneously unfolded in VR."
Nanome’s immediate benefits include understanding the multidimensional nature of the world in which we live. "Shape complementarity (between Novichok and its enzyme target) reinforced by atomic level electronic detail is an essential component of antidote resistant nature of their action," said. Radić. "Its significance is twofold."
"One is to demonstrate an impressive advance in presentation technology," the UC San Diego professor said. "The other is to facilitate understanding of the topic and enable remote participation." He has done this kind of presentation at least twice, so far.
In a public lecture held on July 4th, 2018 in the Zagreb Observatory he first showed 2D structural formulas of suspected Novichok compounds.
"I entered VR while the audience was able to watch my viewpoint in the big screen projection, and then connected in VR with my assistant who was at UCSD in La Jolla at the time," he detailed. "I asked the assistant to open models of the enzyme-bound Novichoks and to hand them to me. He then took over as a presenter while in VR and entered the binding site to show details of interaction to the audience."
His second presentation was in Fall of 2018 at the International meeting in the Czech Republic as a part of a specialized 3D session that he organized and co-chaired. Dr. Radić, who was in VR, had a rendering of selected structural details of his data in stereoscopic 3D (quad buffered 3D stereo) projected in a big screen.
"I opened the structure of the inhibited enzyme, connected to my student at UCSD, and he handed to me a small library of antidote molecules that he was actually working on synthesizing," he explained. "I was able to take each of antidote structures in my hand and demonstrate the way it was supposed to recover activity of the inhibited enzyme and why that recovery was difficult, [which is] due to special properties of bound OP inhibitors."
That Nanome is used in both the process of discovery and education is deeply satisfying to our team. This work would not be possible without the RCSB PDB and the PDB archive.
More information
- Molecule of the Month: Acetylcholinesterase. June 2004, David Goodsell doi:10.2210/rcsb_pdb/mom_2004_6
- U.K. attack shines spotlight on deadly nerve agent developed by Soviet scientists. March 19, 2018, Richard Stone Science doi:10.1126/science.aat6324
- Inside the Virtual Lab — Nanome Power User Zoran Radić. July 20, 2018, Adam Simon, Nanome - Matryx (MTX)


