Data Deposition/Biocuration Services and
Archive Management
Deposition Statistics
In the second quarter of 2025, 5253 experimentally-determined structures were deposited to the PDB archive for a total of 10,536 entries deposited in the year. Data are processed by wwPDB partners RCSB PDB, PDBe, PDBj, and wwPDB Associate Member PDBc.
Of the structures deposited in 2025 so far, 84.3% were deposited with a release status of hold until publication 9.2% were released as soon as annotation of the entry was complete and 6.5% were held until a particular date. 54.9% of these entries were determined by X-ray crystallographic methods 1.3% were determined by NMR methods and 43.3% by 3DEM.
During the same time quarter, 4405 structures were released in the PDB, including 315 SARS-CoV-2 structures. 2762 EMDB maps were released in the archive.
wwPDB News:
Download Snapshots of the PDB Core Archive via HTTPS, SYNC, RSYNC, or FTP
Snapshots of the PDB Core archive (https://files.rcsb.org) can be downloaded via HTTPS, SYNC, RSYNC, or FTP. At RCSB PDB, AWS SYNC is supported instead of RSYNC.
Snapshots have been archived annually since 2005 to provide readily identifiable data sets for research on the PDB archive. Visit wwPDB.org for more information.
Extension to EMDB Accession Codes
To accommodate the rapid growth of EMDB depositions, we are extending the EMDB ID format to six-digits. This change ensures continued archive growth for the coming decade. Visit wwPDB.org for more information.
Announcement: Ligand Expo to Be Retired in 2025

Use the Chemical Sketch Tool at RCSB PDB to create a 2D chemical drawing tool for substructure searching.
RCSB PDB and wwPDB offer robust resources to access chemical and structural information about small molecules within PDB entries.
As such, the legacy Ligand Expo website (ligand-expo.rcsb.org), established nearly 20 years ago as an auxiliary service for small molecules, will be retired in 2025.
Users should transition to RCSB PDB and wwPDB services as soon as possible. Detailed information is available at RCSB PDB.
Electron Density Maps and Coefficient Files
The PDB archive contains two types of data for structures determined by X-ray crystallography. Coordinate files include atomic positions for the final model of the structure and the structure factor file which represents the intensity (or amplitude) for each reflection (X-ray spots) in the diffraction pattern. These two types of data are combined to create electron density maps to represent how well the model fits the data.
wwPDB Validation Reports provide users with detailed information that includes an analysis of the quality of the coordinate model compared with the experimental data. The resulting analysis is available in PDF, XML and CIF formats. For X-ray crystallography structures, 2Fo-Fc and Fo-Fc map coefficient files (weighted amplitudes and phases) containing the data that are used in wwPDB Validation Reports are provided in PDBx/mmCIF format.
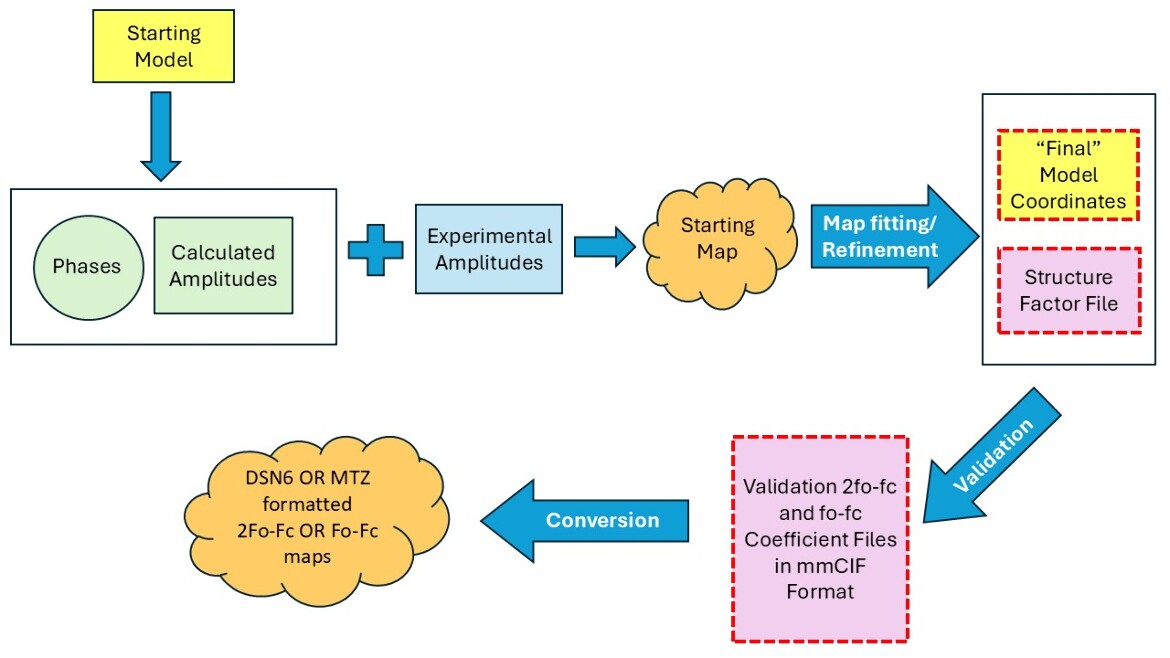
In this diagram, files provided by the PDB are outlined in red, dashed lines.