Data Exploration Services
RCSB.org Statistics
Traffic is tracked using Google Analytics.
| Month | Number of users | Number of sessions |
|---|---|---|
| April 2025 | 789,235 | 1,872,831 |
| May 2025 | 1,061,382 | 2,134,536 |
| June 2025 | 5,713,880 | 6,766,701 |
Explore Integrative Structures at RCSB.org
RCSB.org now offers expanded access to integrative structures. Users can access and explore these complex macromolecular assemblies modeled using integrative methods together with other experimental structures and Computed Structure Models using their favorite RCSB PDB tools.
Integrative structures are determined using a combination of data from traditional techniques such as X-ray crystallography, NMR spectroscopy, and 3D Electron Microscopy (3DEM) along with other experimental approaches like Small Angle Scattering (SAS), Crosslinking Mass Spectrometry, Atomic Force Microscopy, Forster Resonance Energy Transfer, and more. These methods help build structure models of large, dynamic, and heterogeneous biological assemblies that are difficult to resolve with a single technique. Integrative structures are deposited and processed by the wwPDB through PDB-IHM (via OneDep) and archived alongside experimental structures in the PDB archive (announcement). Examples of macromolecular complexes modeled using integrative approaches include the nuclear pore complex, BBSome, and the nucleotide excision repair complex.
RCSB.org offers multiple ways to find integrative structures:
- Keyword or PDB ID searches in the top Basic Search (e.g., “BBSome” or “8ZZE”)
- Advanced Search: Use the Integrative/Hybrid Method Details section to filter by model features (multi-scale, multi-state, ordered states), experimental input types, or related datasets in EMDB, BMRB, PRIDE, and more (Type "IHM" into the attribute box to highlight available options)
- Results Refinement: After any search, filter results by Structure Determination Methodology>integrative
Integrative structures are marked with a distinctive puzzle piece icon, making them easy to identify at a glance in search results.
Structure Summary pages for integrative structures highlight key information about the representative model (see example), and offer download of data files and validation reports.
A user guide offers detailed information on Integrative Structures at RCSB.org.
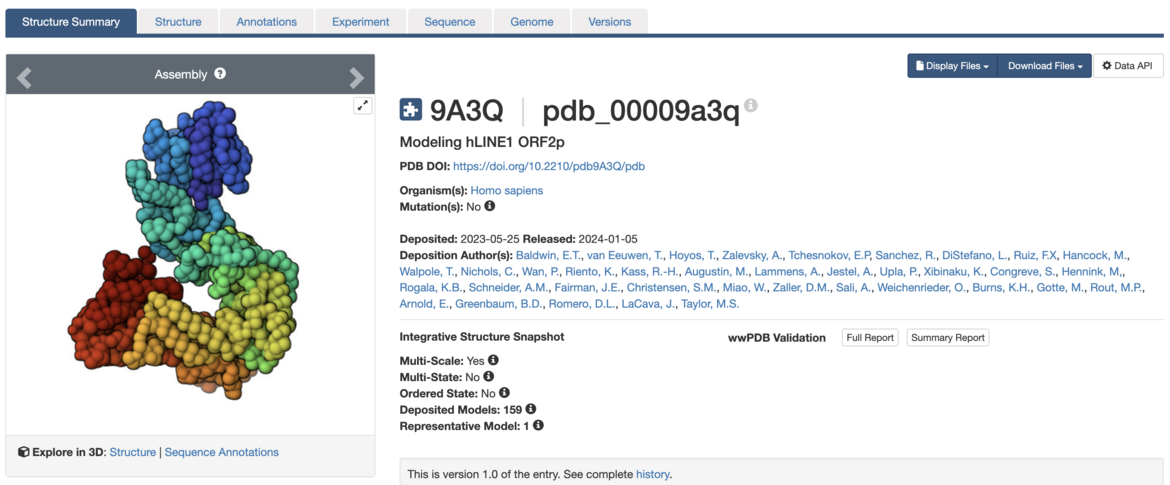
Modeling hLINE1 ORF2p. Visit the Structure Summary to access structure information.
Sequence Annotations Viewer: Geometry Validation
The Sequence Annotations Viewer (help guide |example) maps information from multiple resources and databases onto PDB sequences and structures. Users can select a sequence annotation to focus on the corresponding location on the 3D view of the structure.
Validation scores and geometry validation metrics have been added to the collection of annotations offered.
This visualization enables users to quickly identify regions of interest, structural discrepancies, and potential modeling concerns in an intuitive and interactive manner.
To access this feature, select the Explore in 3D: Sequence Annotations option from the Structure Summary main page or from the Sequence tab (example).
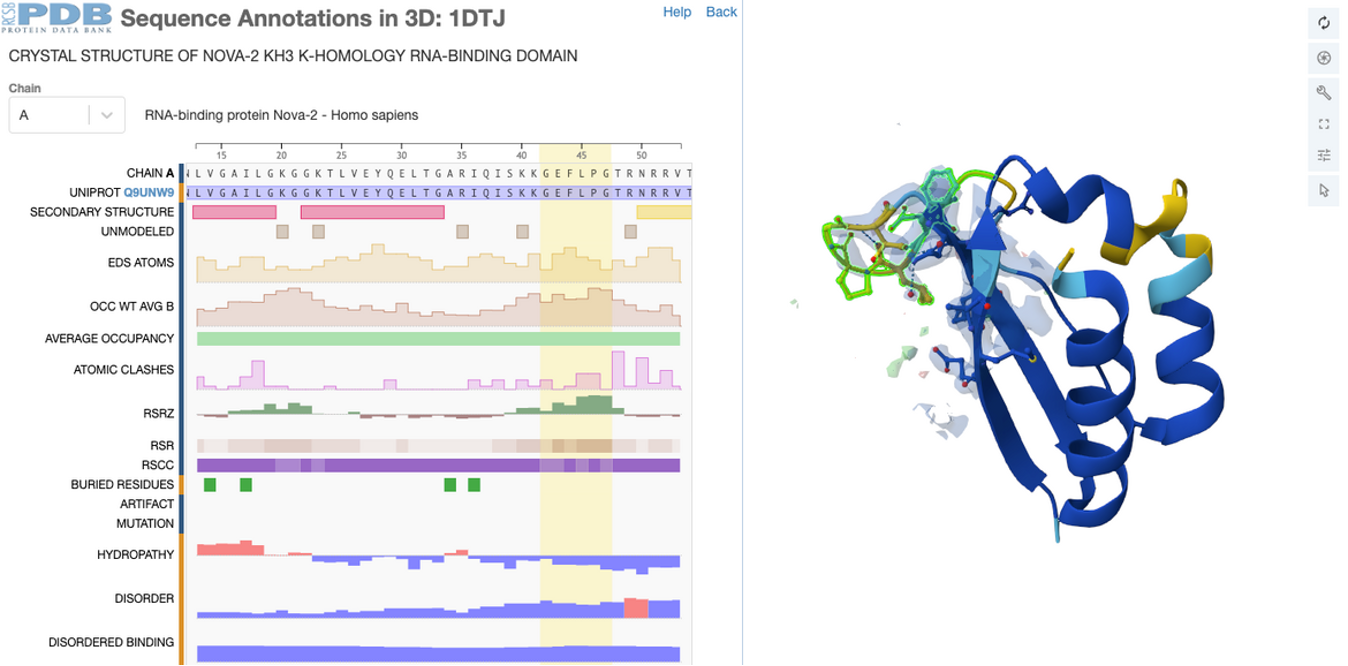
Human RNA-binding protein Nova-2 KH3 domain (PDB ID 1DTJ): most residues have good density except for a few regions as indicated by RSCC and RSRZ values mapped on sequence and RSCC-based confidence levels visualization.
Python Toolkit for Accessing RCSB.org Search and Data APIs

Harness the powerful capabilities of RCSB.org Search and Data API services through the new all-in-one Python toolkit, rcsb-api. This package provides users with streamlined access to the full collection of curated data from the PDB archive together with the rich set of annotations from external resources integrated into RCSB.org, all through a Pythonic interface.
New to APIs? Visit PDB-101 for an introduction.
The Python package can be installed fromPyPI or by downloading the source code on GitHub, which will serve as the central hub for all future development, bug fixes, and project discussions.
The package is organized into distinct modules for each API service—a Search API module and a Data API module—which represent the two main APIs that power RCSB.org. The Search API retrieves PDB IDs that match a given query, while the Data API retrieves data for a given set of PDB IDs. Key features of each module include:
Search API module
- Perform all search types available through the RCSB.org Advanced Search query builder (e.g., full-text, attribute-based, sequence and structure similarity, sequence and structure motif, chemical similarity)
- Use simple Boolean logic to intuitively construct complex or nested queries
- Upload custom structure files for structure similarity searches
- Include computed structure models (CSMs) in search results
- Use faceted queries to aggregate results and gain statistical insights
Data API module
- Retrieve any subset of metadata, features, and/or annotations for a given list of PDB IDs (e.g., experimental method details, structural annotations, binding sites, etc.)
- Easily fetch data for all structures across the archive
- Simplified GraphQL query construction using a Python syntax
Extensive documentation with examples is available, along with a recorded webinar and training materials that introduce RCSB PDB APIs, the rcsb-api Python package, and a series of hands-on notebooks demonstrating usage of the toolkit.
The rcsb-api package—which builds upon previously described work—was developed by Ivana Truong, B.S. (University of Minnesota) and Habiba Morsy, B.S. (Kean University) under the direction of Dennis Piehl, Ph.D. and Brinda Vallat, Ph.D. (RCSB PDB) as part of the Rutgers RISE (Research Intensive Summer Experience and published in the Journal of Molecular Biology:
rcsb-api: Python Toolkit for Streamlining Access to RCSB Protein Data Bank APIs
Dennis W. Piehl, Brinda Vallat, Ivana Truong, Habiba Morsy, Rusham Bhatt, Santiago Blaumann, Pratyoy Biswas, Yana Rose, Sebastian Bittrich, Jose M. Duarte, Joan Segura, Chunxiao Bi, Douglas Myers-Turnbull, Brian P. Hudson, Christine Zardecki, Stephen K. Burley
(2025) Journal of Molecular Biology 437: 168970 doi: 10.1016/j.jmb.2025.168970
Access Validation Reports via API
Users can now programmatically retrieve wwPDB validation reports data via the RCSB PDB Data API. These reports provide critical quality metrics to assess structure quality.
Access overall quality summary, experimental data fit and completeness, and geometric validation (analysis of bond lengths, bond angles, and other stereochemical parameters) for X-ray,EM, and NMR structures.
Key geometry validation metrics have been added to Advanced Search, allowing users to filter structures based on Molprobity angles RMSZ, bonds RMSZ, clashscore, percentage Ramachandran outliers, and percentage rotamer outliers (example).
Introducing Extended PDB IDs at RCSB.org
RCSB.org has introduced features that highlight Extended PDB IDs for all structures to increase visibility and encourage early adoption:
- Search Results & Structure Summary Pages: Extended PDB IDs are displayed, where applicable
- Search: Users can now search for structures using their extended IDs (example)
- Structure Summary URLs: Pages can be accessed using the extended ID, e.g., rcsb.org/structure/pdb_00006UV8
- Data download: Access individual structure data files in PDBx/mmCIF format in a file name that uses extended PDB IDs, e.g., files.rcsb.org/download/pdb_00006UV8.cif
RCSB.org tools and resources will continue to transition to support extended PDB IDs.
About Extended PDB IDs
wwPDB anticipates that all four-character PDB IDs will be exhausted by 2028, after which 12-character PDB IDs will be issued. Entries with extended PDB IDs will not be compatible with the legacy PDB file format and will only be available in PDBx/mmCIF format. wwPDB encourages users to transition to the PDBx/mmCIF format as soon as possible.
The revised PDB accession code format has extended length and prepended “PDB” (e.g., "1abc" will become "pdb_00001abc"). This process will enable text mining detection of PDB entries in the published literature and allow for more informative and transparent delivery of revised data files.
The wwPDB resource portal page (Extended PDB ID With 12 Characters) links to useful resources for this transition, including an FAQ on PDB ID extension, materials to learn more about PDBx/mmCIF format, and links to other PDBx/mmCIF resources and software tools. As the transition phase progresses, more training resources will be added.
Watch a brief presentation by RCSB PDB biocurator Ezra Peisach on Extended PDB IDs.
Additionally, a PDB “beta” archive will be provided during the transition phase in 2026. The directory structure of this “beta” archive will mirror the data organization of the PDB Versioned Archive in the form of https://files-beta.org/pub/pdb/data/entries/two-letter-hash/pdb_accession_code/entry_data_File_names. The two-letter hash will be based on the n-2 and n-3 characters. For example, PDB entry PDB_12345678 will be under /67/. This will maintain consistency with the current PDB archive, where e.g. PDB entry 1abc is under /ab.
All PDB users, including software developers and journal editors, must transition to this format and are encouraged to start using extended IDs and PDBx/mmCIF data files today.
