
|
|
|
|
|
||
Navigating the molecular universe in 3D: Teaching biology students protein structure-function relationships using StarBiochem

Funding for this work was provided in part by the Department of Information Services and Technology at MIT, a Howard Hughes Medical Institute Professorship Grant awarded to Graham Walker, and a grant from the Davis Educational Foundation Grant awarded to John Belcher. The Academic Computing group that participated in this project is now the Software Tools for Academics and Researchers (STAR) group in Office of Educational Innovation and Technology for the Dean of Undergraduate Education at MIT. MELISSA KOSINSKI-COLLINS is now an Assistant Professor of Biology at Brandeis University. Why does a protein do its job in the cell? Because of its shape and chemistry. As biologists, we understand and appreciate the overwhelming knowledge value in these simple statements, but as educators, we battle to try to make the protein structure-function relationship clear to our students. It is easy for us to understand this now, but to get through to our students we need to remember back to a time when we did not understand. How did we originally learn this? The answer is simple: practice. It is clear that a very efficient way to teach to the structure-function relationship comes from letting students view some of the many deposited PDB molecules in a 3D environment. Many of the stereotypically structural "ah-ha" moments come from this type of hands-on interaction with the molecule. Students can not only identify binding pockets and partners, see disease-associated mutations, and observe structural contexts, but they can physically manipulate and, in a sense, control the molecule in real-time. Students of introductory biology need these types of hands-on experiences as well as practice with multiple molecules to really "get" structural biology. Implementing such an interactive yet understandable series of exercises in the average college-level introductory biology course is a daunting task for many reasons. These hurdles include class size, computer and technology access both in the classroom and at home, time devoted to the topic in the syllabus, time involved in creating this type of homework, and the level of understanding of the incoming student. Although there are many freely available software packages that allow the students to explore in 3D, few present the material in a format that makes sense to the average biology student and are simple enough so that the student can use the program outside of the classroom on their own for additional practice. In 2004, a project was begun at MIT to create a new program that filled the pedagogical void left in the world of structural biology. We wanted to create a viewer and a series of exercises that presented structures and functions in the same way we presented them in class, was usable outside of the classroom without staff supervision, and that allowed students many of the freedoms and exploratory options of the research-level PDB viewers. This beta version of this software was named StarBiochem. StarBiochem has one particular option that has become paramount to its success in the context of biology education. In class we invariably introduce protein structure as a build-up of primary, to secondary, to tertiary, to quaternary structures. Most software packages avoid mention of these levels altogether leaving the student to wonder where the levels fit in and how they are related to 3D structure they see on the screen. StarBiochem can open any protein PDB coordinate file and categorize it into these different levels allowing the student to conceptually analyze the 3D structure that they see on their screen. In the examples of hemoglobin and sickle cell anemia, the student is asked to look first at the primary structure change in the molecule, but then to determine at which structural level the disease manifests itself. Using the program as a conceptual guide, the students are asked to understand that the primary structure change from glutamic acid to valine at position 6 does not manifest as a disease until you see a change of intermolecular interaction chemistry in the quaternary structure. StarBiochem was first piloted in an HHMI-sponsored high school field trip at MIT in March 2006. A series of guided exercises led students through an in-depth exploration of proteins with defined structure-function relationships, like sucrose specific porin and hemoglobin. For example, the students were asked to look at the barrel-like structure of porin and reflect on how that shape might be conducive for molecular transport. The students were further asked to investigate the outer chemistry of the molecule and determine how the hydrophobic exterior of the protein might influence the ability of the protein to remain stable in its cellular membrane-bound location. StarBiochem was found to be an effective, and easy-to-use teaching tool in this context and is now used by several of the visiting teachers as a curricular tool in their classroom. The StarBiochem high school initiative is now being further disseminated in MIT and Harvard's Broad Institute Outreach Program. StarBiochem has become an integral part of the introductory biology series at MIT. We have been successful using this program on problem sets and in practice problem assignments. Most of the exercises ask students to visualize a protein that has either been discussed in class or that has a disease connection in StarBiochem. The students explore the 3-dimensional structure of the protein, the stabilizing interactions, and are then asked to relate this information to the biological function of the molecule. The students are given both graded problems and practice problems and have, thus far, been able to use this software on their own at home or in their dorm rooms. We have now moved into the exploration of nucleic acid structures as well. We have developed problems that teach the students how transcription factors and polymerase bind to and function with nucleic acids all based on shape and chemistry. More recently, we have begun to push the boundaries of undergraduate understanding of the protein structure-function relationship even further. Starting in fall of 2007, introductory biology students at Brandeis University will be asked to use StarBiochem to explore the structure of human eye lens protein. They will analyze the shape and chemistry of the molecule in 3D and will then select an amino acid they feel is important to the folding and structure of this protein based solely on their 3D investigation. They will be asked to think about the size and shape of the amino acid, as well as the importance and strength of the interactions in which the amino acid participates. Using site-directed mutagenesis, the students will create their own engineered protein with the ultimate goal of finding and analyzing how or if that residue actually disrupts the structure and the function of the molecule. With the availability of so many structures, let alone so many different molecules, in the PDB, it is our duty as educators to make sure that even the most inexperienced of biology students get opportunities to "see" in the 3D molecular world. We have found both at Brandeis and at MIT that StarBiochem gives students the opportunity to explore the structure-function relationship, and challenges them to go one step further as well. StarBiochem engages students, reinforces classroom concepts, and encourages them to learn as research scientists do; by practice and exploration. StarBiochem is freely available for download at web.mit.edu/star/biochem. Questions about StarBiochem may be sent to kosinski@brandeis.edu. 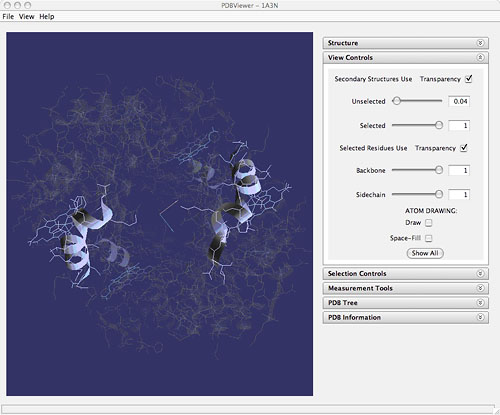
Hemoglobin as viewed using StarBiochem (1a3n: J.R. Tame, B. Vallone (2000) The structures of deoxy human haemoglobin and the mutant Hb Tyrα42His at 120 K. Acta Crystallogr.,Sect.D 56:805-811)
|
||
|
©2007 RCSB PDB |