
|
|
|
|
|
||||||||||||||||||
New Features and Enhancements: Access to Remediation and Pre-remediation Data, Advanced Search, Improved Sequence Details, and Search Result TabsSince the RCSB PDB website and database utilize data from the wwPDB Remediation Project, queries now return more accurate results. New developments in query and reporting features also provide improved access to these data. Access to Remediation and Pre-remediation DataAll data in the PDB archive (ftp://ftp.wwpdb.org) reflects the new features incorporated as part of the wwPDB Remediation Project, including standardized IUPAC nomenclature1 for chemical components. These data have been incorporated into the RCSB PDB website and database to provide improved searching and reporting capabilities. Access to the unremediated data is possible for individual structures and for the entire archive. The left menu of each Structure Summary page provides download options for either remediated or unremediated data in a variety of formats. The Remediation Tab will appear on this page to describe any changes to chain and residue naming conventions made to make the archive more consistent. An example description would be "This structure's single unnamed chain was assigned chain id A". A snapshot of the entire unremediated PDB archive (as of July 31, 2007) is available at ftp.rcsb.org. This archive is not updated. Advanced SearchThe data in the PDB archive offers a wealth of valuable metadata. Advanced Search is a powerful and easy-to-use interface to the underlying search architecture and remediated data. Complex queries are constructed by combining simple "subqueries" chosen from a drop-down list. Users get a feel for the likely success of their search strategy while constructing the search by checking the number of results for each subquery. 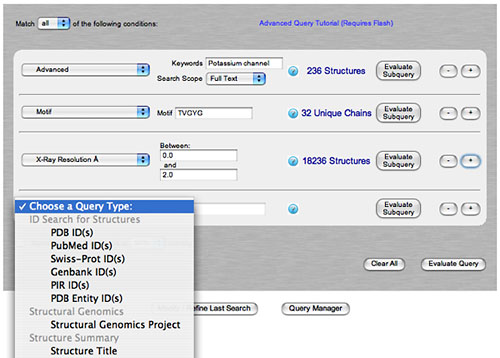
An example of an advanced search which combines searches by keyword, sequence motif, and resolution. Additional queries can be added. A broad range of subqueries is available including sequence searches, Gene Ontology (GO)2 assignments, SCOP3 and CATH4 domain assignments, and author name searches. These subqueries may be combined into a complex query by searching "all" or "any" of the user-specified subqueries. Improved Sequence DetailsThe Sequence Details tab offers a customizable report that displays polymer chain sequences annotated with properties such as domain and secondary structure. This feature utilizes data from the Remediation Project to provide an exact mapping of the structure sequence to the UniProt5 sequence. Annotations from CATH, DSSP,6 PDP,7 and the author-approved secondary structure can be applied to either the sequence in UniProt or in the PDB entry's SEQRES information. The size of the report can be customized for use in presentations. 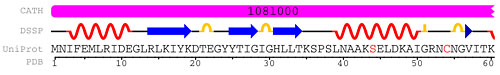
The first 60 residues of T4 lysozyme (108L)8 are mapped to their entries in CATH, DSSP, and UniProt. Mutations in the sequence are shown in red. Search Result TabsKeyword or Advanced Searches will also return different ways of exploring the search results list. Options available from the tabs shown above the default results list include: Citations: The primary citations for all structures have been verified as part of the Remediation Project. This improved mapping between structure and associated reference is reflected in the database. The Citations Tab provides a PubMed-like list of the primary citations for the structures that match a query. Ligand Hits: This tab lists the ligands known to interact with the structures matching the query. For example, a keyword search for "protein kinase" will return all ligands known to bind protein kinases. Linked images, names, IDs, and formulas appear for each ligand. Web Page Hits: Any of the more than 900 curated web pages found at the RCSB PDB website, including Molecule of the Month features, that contain a requested keyword are found on this tab. GO, SCOP, CATH Hits: These tabs provide the hits that map to the Gene Ontology (GO), SCOP and CATH. Entries are returned in a tree browser, which indicates where these structures reside in the respective hierarchies. The SCOP tab, for example, indicates which hits belong to which class of proteins. 
These tabs offer different ways of exploring search results References1 J.L. Markley, A. Bax, Y. Arata, C.W. Hilbers, R. Kaptein, B.D. Sykes, P.E. Wright, and K. Wüthrich (1998) Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the standardization of data bases of protein and nucleic acid structures determined by NMR spectroscopy. Pure & Appl. Chem. 70: 117-142. 2 GO: The Gene Ontology Consortium (2000) Nature Genetics 25:25-29 3 L. Conte, A. Bart, T. Hubbard, S. Brenner, A. Murzin, and C. Chothia (2000) SCOP: a structural classification of proteins database. Nucleic Acids Res. 28(1): 257-259. 4 C.A. Orengo, A.D. Michie, S. Jones, D.T. Jones, M.B. Swindells, and J.M. Thornton (1997) CATH - a hierarchic classification of protein domain structures. Structure. 5: 1093-1108. 5 The UniProt Consortium (2007) The Universal Protein Resource (UniProt). Nucleic Acids Res. 35(Database issue): D193-7. 6 W. Kabsch and C. Sander (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 22: 2577-2637. 7 N. Alexandrov and I. Shindyalov (2003) PDP: protein domain parser. Bioinformatics. 19(3): 429-30. 8 M. Blaber, X.J. Zhang, B.W. Matthews (1993) Structural basis of amino acid alpha helix propensity. Science 260:1637-1640.
WEBSITE STATISTICS
Access statistics for the first quarter of 2007 are given below
for the RCSB PDB website at
www.pdb.org.
|
||||||||||||||||||
|
©2007 RCSB PDB |