DATA
QUERY, REPORTING AND ACCESS
2009 Statistics
2009 Website Statistics
Month |
Unique Visitors |
Visits |
Bandwidth |
January |
160,293 |
355,392 |
634.85 GB |
February |
168,082 |
373,510 |
767.21GB |
March |
189,143 |
434,542 |
966.02 GB |
April |
178,430 |
404,618 |
875.53 GB |
May |
164,084 |
380,493 |
835.61 GB |
June |
145,958 |
356,857 |
1,079.61 GB |
July |
125,701 |
325,381 |
1,064.26 GB |
August |
119,287 |
315,040 |
914.56 GB |
September |
163,204 |
403,770 |
980.24 GB |
October |
195,573 |
471,869 |
1,144.92 GB |
November |
201,967 |
478,142 |
1,324.39 GB |
December |
176,198 |
404,105 |
923.31 GB |
2009 Visitors by City
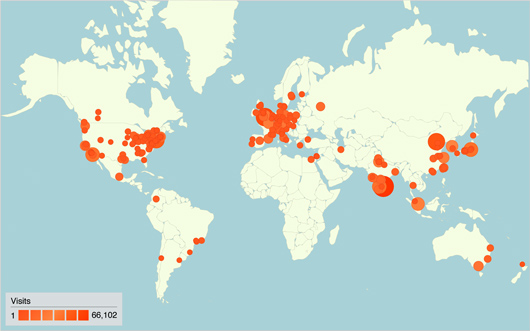
The RCSB PDB website had more than 4 million visits in 2009 by users in more than 15,000 cities. Image created using Google Analytics.
2009 Visitors to the Educational Resources of www.pdb.org
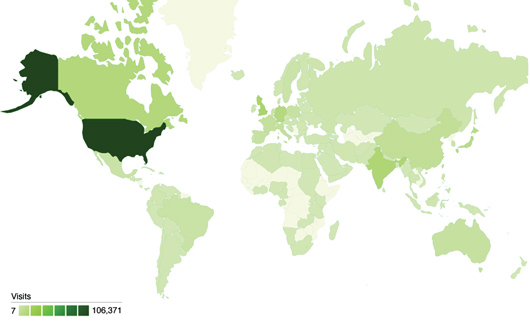
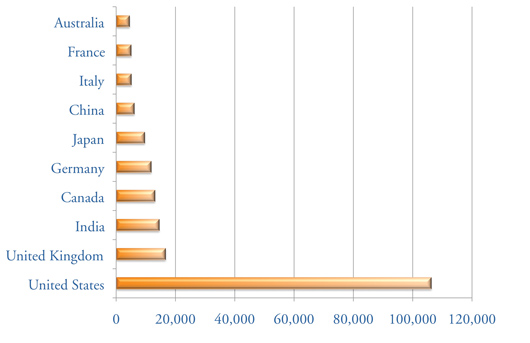
Top: The RCSB PDB's Educational Resources, including Molecule of the Month and Looking at Structures, had more than 300,000 visits by users in 144 countries in 2009. Image created using Google Analytics. Bottom: Top ten visiting countries are shown.
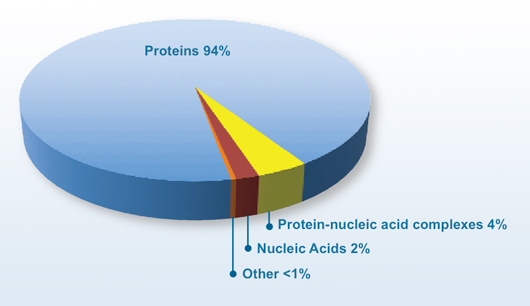
2009 Molecule Type (7448 structures released in 2009)
New and Improved Web Services for Accessing PDB Data
Web Services can help software developers build tools that interact more effectively with PDB data. Instead of storing coordinate files and related data locally, Web Services let software tools interact with the RCSB PDB remotely.
The RCSB PDB supports RESTful services that exchange XML files in response to URL requests. RESTful search services return a list of IDs for Advanced Search and SMILES-based queries. RESTful fetch services return data when given IDs, including PDB entity descriptions, ligand information, third-party annotations for protein chains, and PDB to UniProtKB mappings. SOAP Web Services are also available.
Documentation for accessing these Web Services is available online. Improvements are being made based on community feedback. Please let us know if there are website options that you think should be offered as a web service at info@rcsb.org.
wwPDB FTP Advisory Notice
On November 24, 2009, a few changes were made to the wwPDB FTP directories. Updates were made to the RSYNC script and README files to prompt users to select which wwPDB server should be used (RCSB PDB, PDBe, PDBj).
Sequence cluster data was moved from the wwPDB FTP site to ftp://resources.rcsb.org/sequence/clusters/. This resource contains the results of the weekly clustering of protein chains in the PDB generated by BLASTClust. These clusters are used in the "remove similar sequences" feature on the RCSB PDB website. For more information on this feature, please see the README file and www.rcsb.org/pdb/statistics/clusterStatistics.do
Questions should be sent to the wwPDB at info@wwpdb.org.
New Website Features
A lot of new features have been added to www.pdb.org, including enhancements to browsing query results, generating tabular reports, and viewing large structures. Many of these features are described in this newsletter, but users should always check the  page for detailed descriptions of the latest updates. page for detailed descriptions of the latest updates.
Latest Publications: Access All Articles in Each Update
Available from the left-hand menu, the new Latest Publications
search returns the PubMed articles associated with PDB structures in the most recent update. It includes publications
for newly released structures and for structures whose PubMed abstracts were added to our database in that update.
The results are presented in the Query Results Browser with the "Citations" tab highlighted by default.
The other query results tabs are provided as well, so by clicking on the "Structure Hits"
tab, all structures associated with these publications can be explored. Clicking on the first icon ( ) will download the citations in Medline format.
Improved Tabular Reports
For any search results, users can examine individual PDB entries or review the entire set of structures by creating reports that can be viewed online or downloaded. Recently, this reporting system has been enhanced with improved functionality and navigation, and now offers better support for very large result sets.
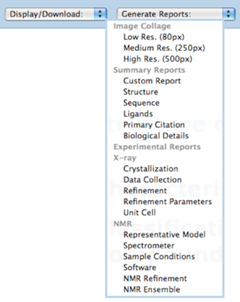
Prepared and customized reports can be generated for search results. • Report Navigation
Reports can be generated for structures matching a query by selecting any of the prepared options available from the pulldown menu or by creating a new, customized report. Instead of presenting everything on a single page, reports are now available on multiple, customizable pages. By default, the first 15 records sorted by PDB ID are shown, with an option to list more records per page.
The entire table can be sorted by clicking on column headers. Column widths can be resized by dragging the line between two columns. Within the reports themselves, PDB IDs link to that entry's Structure Summary page, PubMed IDs to the abstract, and Ligand IDs to a Ligand Summary page.
• Exporting Reports
Tables can be exported in three formats: Excel 97-2003, Excel 2007 or newer, and or Comma Separated Value (CSV) format (recommended for extra large data sets that may surpass size limitations in Excel).
The Excel spreadsheets are formatted with customized column width, text wrapping, alignment, and hyperlinks on selected columns.
• Generating Large Reports
Reports can now be generated for extremely large sets of data. For example, a report displaying the X-ray Refinement Details (Rvalue, Rfree, and Resolution) can be made for all crystal structures in the PDB.
To generate this report, select PDB Statistics at top right of the RCSB PDB website, and click on the first link for Summary Table of Released Entries.
Selecting the number for X-RAY / Total will return the more than 53000 structures that match this query. From the navigation bar, click on Generate Reports and select Experimental Reports/X-ray/ Refinement. This report can be browsed, formatted, and downloaded as described above.
Other large reports, such as the summary reports for Biological Details and Sequence, can be easily generated for all structures in the PDB.
Improved Display of Large Structures
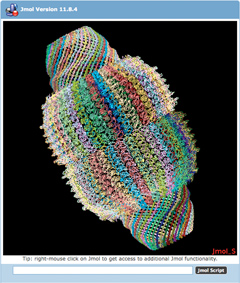 The vault structure represented by entries 2zuo, 2zva, 2zv5 can now be viewed in
a single Jmol viewer at the RCSB PDB.Images and Jmol1 displays on the RCSB PDB website now show the complete biological assembly for all structures–even for proteins that are split across multiple PDB coordinate files. The vault structure represented by entries 2zuo, 2zva, 2zv5 can now be viewed in
a single Jmol viewer at the RCSB PDB.Images and Jmol1 displays on the RCSB PDB website now show the complete biological assembly for all structures–even for proteins that are split across multiple PDB coordinate files.
A number of structures in the PDB archive are so large that the historical limitations of the PDB file format (maximum of 99999 atoms and 62 unique chains) require them to be split across multiple PDB coordinate files. Examples include extremely large ribosome complexes (e.g., 1gix, 1giy), and structures that contain a very large number of atoms or chains, such as the vault protein (e.g., 2zuo, 2zva, 2zv5).
These structures are identified on Structure Summary pages in the new "Split Entry" box, which lists and links the PDB IDs of all entries that make up the composite structure. A link is provided to easily download all of the related coordinate files.
Images on the Structure Summary page now illustrate the full composite structure, rather than what is found in each individual PDB coordinate file. The image on the Structure Summary page for 1gix, for example, shows the full ribosome structure composed of entries 1gix and 1giy. For all Structure Summary images, the forward and backward buttons can be used to toggle between the asymmetric unit and the biological assembly (or multiple biological assemblies, if applicable) for the full structure.
The biological assembly and asymmetric unit of the composite structure can be launched in Jmol when viewing the corresponding static image. The display of such large structures in Jmol is possible by loading PDB files limited to CA and P atoms, and by using the Jmol load files command.
Use Widgets to Embed RCSB PDB Features on Your Website
 These widgets link directly to the RCSB PDB website so users don't have to download any files. The Molecule of the Month widget can be used to link to a particular feature, or it can be set to be automatically updated when new features are published.Molecule of the Month images and text, static images and interactive views for any PDB structure, and RCSB PDB searches based on ID and keyword can be embedded into any website using the RCSB PDB's web widgets. These small bits of code can be customized to display and link to RCSB PDB features. These widgets link directly to the RCSB PDB website so users don't have to download any files. The Molecule of the Month widget can be used to link to a particular feature, or it can be set to be automatically updated when new features are published.Molecule of the Month images and text, static images and interactive views for any PDB structure, and RCSB PDB searches based on ID and keyword can be embedded into any website using the RCSB PDB's web widgets. These small bits of code can be customized to display and link to RCSB PDB features.
• The RCSB PDB Molecule of the Month widget embeds an image from the feature and links to the entire article. The widget can be customized to specify width, colors, amount of text shown, and molecule.
• The Tag Library can be used to embed an image that links to the corresponding Structure Summary page; provide a menu of links to a particular entry's Jmol view, Structure Summary Page, and PDB file; and provide a link that performs a current keyword or author query.
• The Image Library widget embeds an image of a structure based on PDB ID.
These widgets bring RCSB PDB functionality to any resource. For more information and other widgets, see the Widgets page linked from the Tools section of the RCSB PDB's left menu.
Improved Advanced Search Interface and Help
A new Advanced Search interface helps users build complex queries in a more intuitive manner. Search criteria are more clearly defined, and the overall design makes it easy to add and remove search parameters.
Advanced Search is a tool that can be used to combine a variety of simple searches into a single query, such as: Which proteins have ligands (but not any nucleic acids)? What protein-serine/threonine kinases were released in 2008? How many structures with resolution less than 2.0 Å contain a given sequence motif?
Advanced Search has been enhanced with a new intuitive help system that will eventually replace the current robohelp system.
The Advanced Search help system displays help, query definitions, and examples based upon what is shown on the screen. For
example, selecting at the first Advanced Search page will
return an overview of the query feature, while selecting
when looking at the window for Secondary Structure Content returns context-related search criteria and examples. By
default, the new help system opens in a shadow box interface, but can also be viewed in a separate window by clicking
on the pop-up link.
1. Jmol: an open-source Java viewer for chemical structures in 3D. jmol.sourceforge.net
|







