DATA
QUERY, REPORTING AND ACCESS
2011 RCSB PDB Website Statistics
Month |
Unique Visitors |
Visits |
Bandwidth |
January |
210986 |
522651 |
982.26 GB |
February |
213862 |
526896 |
1114.34 GB |
March |
239640 |
598545 |
1004.99 GB |
April |
230825 |
557911 |
763.04 GB |
May |
233123 |
561880 |
814.08 GB |
June |
206456 |
507798 |
620.89 GB |
July |
187344 |
470491 |
589.52 GB |
August |
189689 |
467477 |
642.00 GB |
September |
242775 |
567725 |
683.56 GB |
October |
274335 |
646682 |
800.58 GB |
November |
280058 |
660068 |
945.80 GB |
December |
233065 |
547841 |
673.20 GB |
Latest Website Release
New and enhanced features have been added to the RCSB PDB website, including:
Surface of PBCV-1 virus capsid with 5040 chains; one of the largest assemblies in the archive (PDB ID 1m4x). Image created using Protein Workshop's new Surfaces option.
- Visualizing molecular surfaces. Protein Workshop now supports molecular surfaces to aid in the display of quaternary structure, protein-protein interactions, and binding sites. The program is one of the molecular viewing options available from every Structure Summary page.
- Exploring ligands. Improved Jmol visualization, information about subcomponents, and links to DrugBank are some of the features added to Ligand Summary pages. Additionally, binding affinity data from PDBbind (www.pdbbind-cn.org) have been added to the External Ligand Annotations widget on Structure Summary pages.
- Access pre-released sequences via Web Services. The RCSB PDB supports RESTful Web Services, which can be used to help software developers access data more easily. A new service for accessing pre-released sequences in FASTA format has been added; information about all possible services is available at http://bit.ly/tiqkzs.
The What's New page at http://bit.ly/rJPaA2 has complete descriptions of all the new features.
Looking for Ligands
All residues and small molecule components found in PDB entries, including standard and modified amino acids/nucleotides, small molecule ligands, ions, and solvent molecules, are described in the wwPDB's Chemical Component Dictionary.
The information stored in this dictionary can be easily searched using any of these query options:
- Simple top bar searching (with autocomplete suggestions). Click on the Ligand icon (located above the top search box) to limit your search. Enter a ligand name or the 3-character chemical component ID.
- Chemical Component Search. This powerful form can be accessed by selecting the top bar ligand icon and then [ additional ligand options ]. Launch the chemical structure editor to draw a structure, or paste in a SMILES or SMARTS string, and then perform a substructure, exact, similarity, or superstructure search. In addition, ligands can be searched by name, identifier, formula, and molecular weight. A tutorial and screencast demonstrating the chemical structure search is available at http://bit.ly/ruxFLI.
- Advanced Search. Supports the types of searches mentioned above, plus options to query by chemical component type, binding affinity, sub-component, and more. These searches can be combined and even amended with searches for the associated structures.
These searches will return a Ligand Summary page that contains an overview of the chemical component, 2D and 3D images, links to other resources, and links to related ligands and PDB entries.
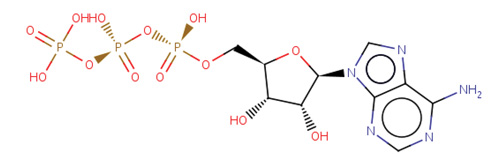 Image of ATP from the Ligand Summary page Image of ATP from the Ligand Summary page
Search the PDB for Homo- and Hetero-multimer Structures
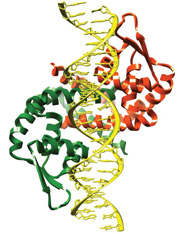 Protein homodimer bound to DNA as seen in PDB ID 3q5f. Protein homodimer bound to DNA as seen in PDB ID 3q5f.
The following Advanced Search options (listed under Structure Features) can be combined to find homo- and hetero-multimer structures in the PDB:
- Number of Chains (Biological Assembly): specifies the number of polymeric chains in a multimer
- Number of Entities: limits the number of unique polymeric units (protein, DNA, or RNA) in a multimer
- Structure Features/Macromolecule Type: selects the type(s) of macromolecule to be included in the multimer (protein, DNA, RNA, and/or DNA/RNA hybrid).
Detailed examples of these types of searches can be found in the RCSB PDB news at http://bit.ly/tdNlan.
Customize Your RCSB PDB Homepage
The RCSB PDB homepage is comprised of web widgets that can be moved around, minimized, or hidden so users can create a website that reflects their interests.
Frequently-used features can be moved to the top, while less popular items can be removed from the page or collapsed by selecting Hide. Widget boxes with an arrow can be dragged up and down on the page. Select the Customize This Page button from the left menu to choose which sections are displayed.
Options in the default view include:
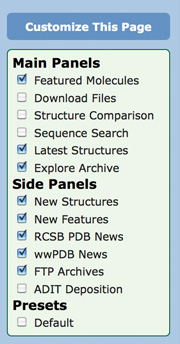 Customize the RCSB PDB homepage with different widgets. Customize the RCSB PDB homepage with different widgets.
- Featured Molecules: Read the latest Molecule of the Month and the
PSI SBKB's Featured System.
- Latest Structures: Scrolls through a slideshow of individual entries,
with links to the related abstract, Structure Summary page, and
Jmol view.
- Explore Archive: Tour the PDB archive by "drilling-down" on
significant properties of structures like Organism and Polymer Type.
This widget also gives a quick statistical overview of the archive.
- New Structures: Links to the latest release, structure papers included in
a release, and to the Unreleased Entry Search.
- New Features: Scrolls through the latest website features and
improvements with links to detailed descriptions.
- RCSB PDB News: Read the latest updates and highlights,
Newsletter and more.
- wwPDB News: Updates from wwPDB.org are linked in this widget.
- FTP Archives: Links to the current FTP archive and to archival
snapshots.
Widgets not included in the default view that can be added are:
- Download Files: Provides a form on the home page to easily
download any number of structures in a variety of formats.
- Structure Comparison: Run pairwise structural and sequence
alignments.
- Sequence Search: Enter a PDB ID or sequence to query for similar
sequences.
- ADIT Deposition: Provides a quick interface to the RCSB PDB
ADIT deposition services. Users can start a new deposition, or
continue an existing session.
Select a combination of widgets to display, and then move the widgets around to customize the display.
Structure Summary pages and query result pages can be similarly customized.
Use the Top Bar Suggestion Box for Quick and Precise Searching
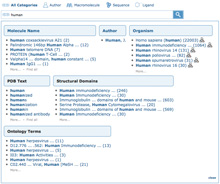 Pop-up box showing suggestions for human. (Click to enlage) Pop-up box showing suggestions for human. (Click to enlage)
The top search bar has been redesigned to help users easily and intuitively create precise searches.
Typing just a few characters launches an interactive pop-up box with suggestions of common PDB search terms, organized in different categories.
These categories return more precise results than simple text searches. For example, entering the word human presents several options organized by category (such as Molecule Name, Author, Organism). Suggestions include the number of results and link to the set of matching structures. Results for organism Homo sapiens (human) will not include entries of author Human, J.
The redesigned top search can be also be limited to quick searches on Author, Macromolecule name, Sequence, or Ligand by selecting the related icon.
The order of results of a PDB text search or a sequence search is now based on the relevance of the term (for a text search) or the alignment score (for a sequence search). Search results can be further refined using Advanced Search or by drilling down through the pie charts organized by major categories.
EMDB to Join the PDB Archive
The EM Data Bank (EMDB), the primary archive for
experimentally-determined maps obtained using
3D electron microscopy methods, will join the PDB archive (ftp://ftp.wwpdb.org) on March 7, 2012. For more information, see wwpdb.org/em.
Time-stamped Copies of the PDB Archive
A snapshot of the PDB archive (ftp.wwpdb.org) as of January 2, 2012 has been added to ftp://snapshots.wwpdb.org/. Snapshots have been archived annually since January 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20120102 includes the 78,237 experimentally-determined coordinate files and related experimental data that were available at that time. Coordinate data are available in PDB, mmCIF, and XML formats. The date and time stamp of each file indicates the last time the file was modified.
The script at ftp://snapshots.wwpdb.org/rsyncSnapshots.sh may be used to make a local copy of a snapshot or sections of the snapshot.
|

 Customize the RCSB PDB homepage with different widgets.
Customize the RCSB PDB homepage with different widgets.


