Data Query, Reporting and Access
Website Statistics
Access statistics for the first quarter of 2013 are shown.
Month |
Unique Visitor |
Visits |
Bandwidth |
January |
308327 |
703069 |
1785.92 GB |
February |
317729 |
683177 |
1866.81 GB |
March |
346548 |
765109 |
1529.32 GB |
Access Drugs and Drug Targets in the PDB
New features for exploring drugs found in PDB entries were recently added to the website.
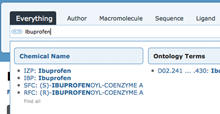
Auto-complete suggestions of stereoisomers when searching for ibuprofen.
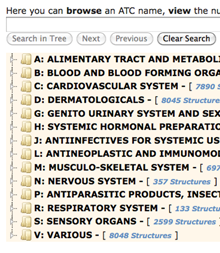
Enter search terms or browse through categories to find PDB structures in a given Anatomical Therapeutic Chemical class.
DrugBank Widget A new Drug Info widget available from Ligand Summary pages lists the corresponding data from DrugBank (when available). The widget contains DrugBank ID, drug name, groups, brand name, and more, with links to the corresponding data at DrugBank. A sequence search based on the drug target sequence can be launched from this widget. For an example, see the Ligand Summary page for BP (Ibuprofen).
Browse Database using ATC The WHO Collaborating Centre for Drug Statistics Methodology's Anatomical Therapeutic Chemical (ATC) classification system organizes drugs into five levels according to the organ or system on which they act and/or their therapeutic and chemical characteristics. The RCSB PDB database can now be browsed using this system. Select the ATC tab from the Browse Database interface to navigate through the drug classification hierarchy, view the number of associated PDB structures, and access the related entries.
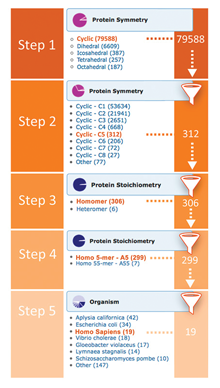
Distribution of PDB Data by Protein Stoichiometry and Symmetry
Distribution drill-downs for protein stoichiometry and symmetry (at 95% sequence identity threshold) have been added to the Explore Archive widget on the home page. The stoichiometry and symmetry information comes from the first biological assembly associated with the entry.
The drill-downs can be applied successively. For example to find C5 symmetric homo-pentameric human proteins one would use the following sequence of drill-downs:
Step 1: Protein Symmetry Cyclic
Step 2: Protein Symmetry C5
Step 3: Protein Stoichiometry Homomer
Step 4: Protein Stoichiometry A5
Step 5: Organism Homo sapiens
Drill-downs, including options for stoichiometry or symmetry, can also be used to further refine search results from the query result browser page.
Advanced Search can also be used to search for structures based on protein stoichiometry and symmetry.
3D Visualization of Protein Symmetry
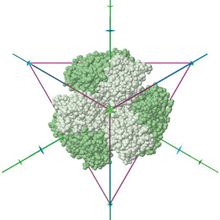
To create this image, select the "View in 3D" link for PDB ID 1G63, and then select Jmol options polyhedron: on; axes: on; style: CPK; color: symmetry; and deselect background: black.
To facilitate the exploration of symmetry, several options are available from the 3D Jmol views of structure:
Default Orientations: Users can toggle between canonical views of structure that highlight symmetry: sides and back, and along unique n-fold symmetry axes.
Symmetry axes and polyhedra. Symmetry axes and symbols representing folds (dyad for 2-fold, triad for 3-fold axis, or in general a polygon for n-fold axis) can be displayed with a structure. Users can select to view the protein enclosed in a polyhedron that matches its symmetry.
Colored by symmetry or subunit. Structures can be colored by symmetry (see the list of color schemes used), sequence (subunits with >= 95% sequence identity shown in the same color), or by subunit.
Pseudosymmetry. The default view shows symmetry based on a 95% sequence identity threshold. Since some structures will display pseudosymmetry when using a 30% sequence identity threshold, an option lets users toggle between the two options.
To access Jmol on the RCSB PDB site, select the "View in 3D" link on any entry's Structure Summary page. Protein symmetry is calculated for all entries containing at least one protein chain, including asymmetric units and all biological assemblies (except for entries split among several PDB files due to their size).
Quick Tour of Search Results
A new video demonstrates the different options available for exploring search results at the RCSB PDB website.
Search, View Annotations, and Visualize Peptide-like Antibiotic and Inhibitor Molecules

Autosuggestions from a Simple Search for actinomycin
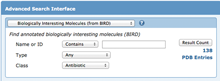
Advanced Search options for querying BIRD
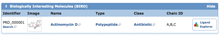
BIRD widget from PDB ID 1a7y
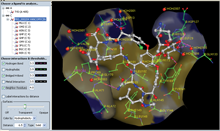
Ligand Explorer view of Vancomycin PRD_000204.
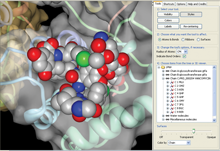
Protein Workshop view of vancomycin PRD_000204 as seen in PDB ID 1pnv.
New RCSB PDB website features utilize BIRD data to provide improved searching and visualization options for these molecules:
Search BIRD Molecules
Simple Search: Biologically interesting Molecules from BIRD can be searched by typing a name (vancomycin), a BIRD ID (PRD_000204), type (glycopeptide), or class (antibiotic) in the top search bar. Suggestions will appear under the BIRD Molecules category.Advanced Search: Biologically interesting Molecules from BIRD can also be searched from the advanced search menu. Search by text/name, BIRD type (structural classification of the entity), or BIRD class (broad definition of the entity function).
View Annotations from BIRD: The BIRD widget on an entry's Structure Summary page will display the ID, image, name, type, class, and chain location for any such molecules in the entry. Ligand Explorer can be launched to view the molecule and binding site in 3D.
As with other Structure Summary page features, the examples displayed (ID, name, type, and class) can be used to find other PDB entries with the same characteristics.
Visualize BIRD Molecules
Ligand Explorer: Ligand Explorer can be launched from the BIRD Widget to visualize these molecules and their binding sites. The program can center on other molecules by clicking on a name or identifier from the left hand menu. Intermolecular interactions such as hydrogen bond or hydrophobic interactions and a binding site surface can be turned on for the active ligand.Protein Workshop: In Protein Workshop, BIRD molecules are listed in the right hand menu and can be manipulated as with any other macromolecule chain or ligand. Protein Workshop can be launched from any entry's Structure Summary page.
wwPDB News
Time-stamped Copies of the PDB Archive
A snapshot of the PDB archive (ftp.wwpdb.org) as of January 1, 2013 has been added to ftp://snapshots.wwpdb.org/. Snapshots have been archived annually since January 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20130101 includes the 87,090 experimentally-determined coordinate files and related experimental data that were available at that time. Coordinate data are available in PDB, mmCIF, and XML formats. The date and time stamp of each file indicates the last time the file was modified.
The script at ftp://snapshots.wwpdb.org/rsyncSnapshots.sh may be used to make a local copy of a snapshot or sections of the snapshot.