
Dr. Josh Beckham is an Assistant Director and Research Educator with the Freshman Research Initiative in the College of Natural Sciences at The University of Texas at Austin. He teaches computational and molecular biology techniques in the context of drug discovery for infectious diseases while fostering the development of the program and promoting external relations.

Student researchers in the Virtual Drug Screening stream
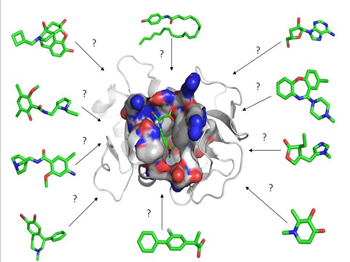
Figure 1: A representation of an active site with small molecule
compounds arrayed around the protein to be docked
and scored by a virtual screening program.
Made in PyMol®.
Alyssa and Melissa pipetting in lab
Joshua presenting his research on a virulence
phosphatase
in Mycobacterium tuberculosis at
the NCUR 2012 conference.

Kathryn presenting her work on a fatty acid synthesis
enzyme (FabI) at the undergraduate poster session at UT-Austin
Imagine the impact of being able to do research in your first years of college. Would it have changed your career path? Would you have understood your coursework better and appreciated your undergraduate experience more? The value of research to an education in the sciences has been documented extensively 1,2 The challenge is being able to implement research on a large scale in a meaningful way for students of all backgrounds.
Students at The University of Texas at Austin that are in the FRI (Freshman Research Initiative) program have this opportunity. The FRI is a novel approach that brings the missions of education and research together. Initiated in 2005 by NSF funding and supported in part by the Howard Hughes Medical Institute (HHMI), the program involves over 800 freshmen and sophomores in research projects under both a tenured faculty member and a non-tenure track 'Research Educator' in courses that we call 'Streams'.