 Users can quickly find all annotated membrane proteins in the PDB by entering "membrane proteins" in the top bar simple search and selecting the "Retrieve Membrane Proteins" option.
Users can quickly find all annotated membrane proteins in the PDB by entering "membrane proteins" in the top bar simple search and selecting the "Retrieve Membrane Proteins" option.
The Membrane Protein Browser and the Membrane Proteins drill-down tool from the home page and search results can be used to investigate specific membrane protein classifications and access the corresponding structures.
Membrane protein annotations for each entry appear in the Search Results and individual Structure Summary pages (example: 2rh1).
Transmembrane proteins in the PDB are identified using the mpstruc database (Stephen White, UC Irvine), sequence clustering, and data derived from UniProt as described at rcsb.org.

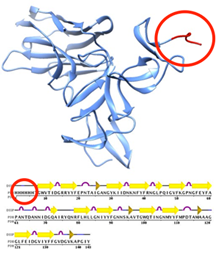
Entry 4bnx contains a His6 tag (circled).
Protein tags are peptide sequences attached to proteins for various experimental reasons, particularly for purification. Advanced Search can be used to search for entries with these protein purification tags.
To find these structures, select the Sequence Features > Sequence Motif option and enter an exact sequence or a sequence pattern expressed by regular expression syntax. Regular expressions can be used to define complex sequence patterns.
Entering ^HHHHHH will search for a 6 residue long N-terminal polyhistidine-tag (His6 tag), while a search for HHHHHH will find any sequence with a (His)6 pattern. This query looks at the sequence originally studied, and will find entries where an His6 tag appears completely, in part, or not at all in the coordinates.
A detailed description of using the Advanced Search sequence motif option is available at rcsb.org.

The biotin binding site in PDB entry 1stp. To access this view, select the 'Ligands' tab and click on 'View Pocket in Jmol'.
Options for visualizing binding sites have been added to the Jmol/JSmol views available at rcsb.org.
From a Structure Summary page (example: 1stp), select the option for "3D View" listed under the entry's image (or from the 3D View tab).
Options for viewing domains and ligands are at the bottom of the page.
Select the Ligand tab and then the link to "View Pocket in Jmol" to access this new feature. Residues are displayed in contact with the selected ligand. A cropped ligand van der Waals surface will appear that is color coded by the proximity to the van der Waals surface of the binding site (red: close contact, blue: far contact).
Thanks to Robert Hanson (St. Olaf College), the current principal Jmol/JSmol developer, for implementing this feature during his visit to the RCSB PDB.
A snapshot of the PDB archive (ftp://ftp.wwpdb.org) as of January 2, 2014 has been added to ftp://snapshots.wwpdb.org/. Snapshots have been archived annually since January 2005 to provide readily identifiable data sets for research on the PDB archive.
The directory 20140102 includes the 96,692 experimentally-determined coordinate files and related experimental data that were available at that time. Coordinate data are available in PDB, mmCIF, and XML formats. The date and time stamp of each file indicates the last time the file was modified.
The script at ftp://snapshots.wwpdb.org/rsyncSnapshots.sh may be used to make a local copy of a snapshot or sections of the snapshot.
| Month | Unique Visitors | Visits | Bandwidth |
|---|---|---|---|
| January 2014 | 269,913 | 635,303 | 1888.15 GB |
| February 2014 | 284,320 | 642,443 | 1897.20 GB |
| March 2014 | 315,871 | 743,752 | 2151.62 GB |