Data Deposition/Biocuration Services and
Archive Management
PDB Reaches a New Milestone: 200,000+ Entries

On January 10, 2023, the PDB archive holdings passed a milestone 200,000 structure entries. The archive passed 150,000 structures in 2019 and 100,000 structures in 2014.
Established in 1971, this central, public archive has reached this critical milestone thanks to the efforts of structural biologists throughout the world who contribute their experimentally-determined protein and nucleic acid structure data.
wwPDB data centers support online access to three-dimensional structures of biological macromolecules that help researchers understand many facets of biomedicine, agriculture, and ecology, from protein synthesis to health and disease to biological energy. Many milestones have been reached since the archive released the 100,000th structure in 2014. PDB data have been seminal in understanding SARS-CoV-2, and provided the foundation for the development of AI/ML techniques for predicting protein structure. The 50th anniversary of the PDB was celebrated throughout 2021.
Today, the archive is quite large, containing more than 3,000,000 files related to these PDB entries that require more than 1086 Gbytes of storage. PDB structures contain more than 1.8 billion non-hydrogen atoms.
Deposition Statistics
In the first quarter of 2023, 3896 experimentally-determined structures were deposited to the PDB archive for a total of 3896 entries deposited in the year. Data are processed by wwPDB partners RCSB PDB, PDBe, PDBj and PDBc.
Of the structures deposited in 2023 so far, 86.1% were deposited with a release status of hold until publication, 7.1% were released as soon as annotation of the entry was complete, and 6.8% were held until a particular date. 62.8% of these entries were determined by X-ray crystallographic methods. 1.9% were determined by NMR methods and 35.0% by 3DEM.
During the same time quarter, 3399 structures were released in the PDB, including 138 SARS-CoV-2 structures. 1613 EMDB maps were released in the archive.
Access Depositions Using ORCiD
PDB contact authors can now use ORCiDs to authenticate OneDep access. This authentication method allows each contact author to login to OneDep without the need for password sharing to view and access corresponding depositions.
OneDep login using a deposition ID and password is still possible, but will only provide access to the specific deposition.
Using ORCiD with OneDep returns a summary table of the entries in which the ORCiD has been provided for the contact author. Users can further access each of their entries’ deposition interfaces without the need to login again using a deposition ID or password.
Providing ORCiDs for OneDep contact authors has been mandatory since 2018.
After using the ORCiD login, this OneDep panel will display all available depositions. Visit wwPDB.org for details.
Length of CCD and PDB IDs to Change
PDB entries with extended CCD or PDB IDs will be distributed in PDBx/mmCIF format only.
wwPDB, in collaboration with the PDBx/mmCIF Working Group, has set plans to extend the length of accession codes (IDs) for PDB and Chemical Component Dictionary (CCD) entries in the future. PDB entries containing these extended IDs will not be supported by the legacy PDB file format. (see previous announcement)
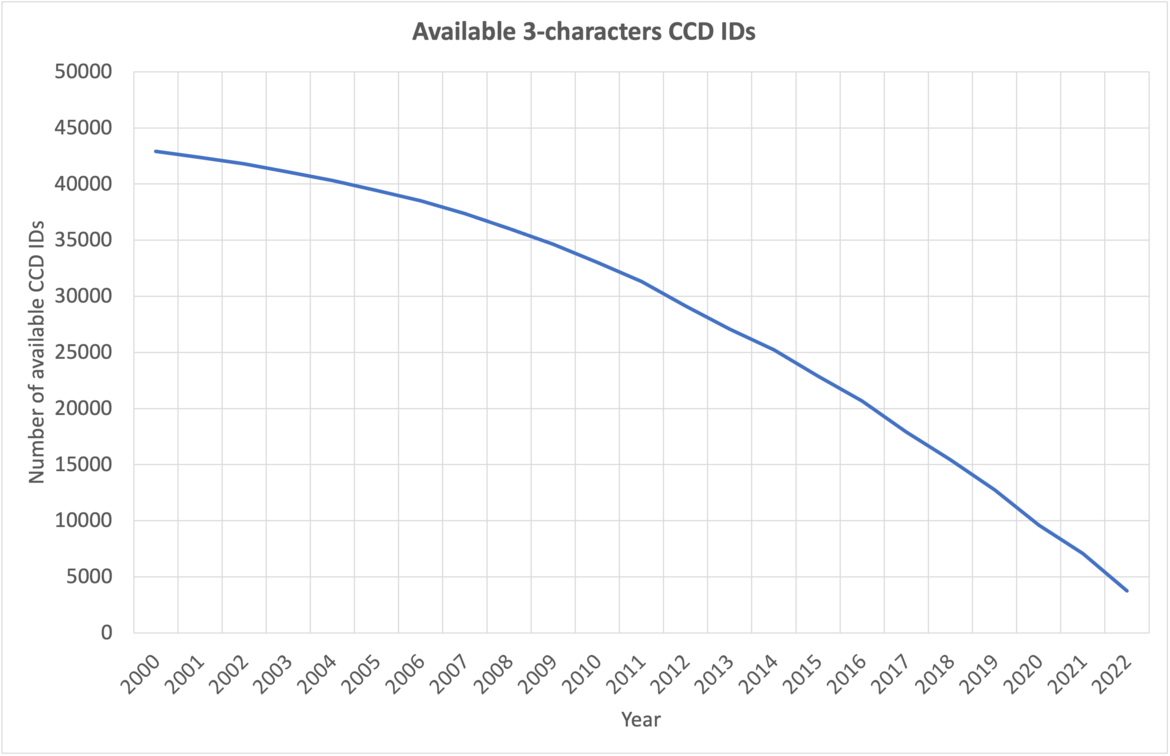
The number of available 3-character CCD IDs annually.
CCD entries are currently identified by unique three-character alphanumeric IDs. At current growth rates, we anticipate running out of three-character IDs before 2024. After this point, the wwPDB will issue five-character alphanumeric accession codes for CCD IDs in the OneDep system. To avoid confusion with current four-character PDB IDs, four-character codes will not be used. Owing to limitations of the legacy PDB file format, PDB entries containing the new five character ID codes will only be distributed in PDBx/mmCIF format.
In addition, wwPDB has reserved a set of CCD IDs: 01 - 99, DRG, INH, LIG that will never be used in the PDB. These reserved codes can be used for new ligands during structure determination so that they can be identified as new upon deposition and added to the CCD during biocuration.
PDB ID extension
wwPDB will be extending PDB ID length to eight characters prefixed by ‘pdb’, e.g., pdb_00001abc. Each PDB entry has a corresponding Digital Object Identifier (DOI), often required for manuscript submission to journals and described in publications by the structure authors. Extended PDB IDs and corresponding PDB DOIs have been included in the PDBx/mmCIF formatted atomic coordinate files for all new and re-released entries since August 2021.
Resources
wwPDB is asking users and software developers to review their code and remove any current limitations on PDB and CCD ID lengths, and to enable use of PDBx/mmCIF format files. Example files with extended PDB and/or CCD IDs are available via github to assist code revisions, see https://github.com/wwPDB/extended-wwPDB-identifier-examples. To learn about PDBx/mmCIF, please visit https://mmcif.wwpdb.org/.
PDB NextGen Archive Prototype Now Available
A prototype of a next generation archive repository for the PDB is now available. The archive, called “NextGen”, hosts structural model files in PDBx/mmCIF and PDBML formats at files-nextgen.wwpdb.org. This enriched PDB archive provides annotation from external database resources in the metadata in addition to the content provided in the structure model files in the PDB main archive at files.wwpdb.org.
This prototype provides sequence annotation from external resources such as UniProt, SCOP2 and Pfam at atom, residue, and chain levels. This mapping information is derived from the Structure Integration with Function, Taxonomy and Sequence (SIFTS) project (https://www.ebi.ac.uk/pdbe/docs/sifts/), a service developed and maintained by the PDBe and UniProt teams at EMBL-EBI. Sequence mappings are provided in _pdbx_sifts_unp_segments and _pdbx_sifts_xref_db_segments categories for each segment, _pdbx_sifts_xref_db at residue level, and _atom_site at atom level.