Education Corner
The UGA Biotech Boot Camp:
Introducing Georgia High School Teachers to Biotechnology and the PDB
by John Rose, Wendy Dustman and Julie Kittleson

Dr. John Rose is an Associate Professor in the Department of Biochemistry & Molecular Biology at the University of Georgia, where he teaches Introduction to Biochemistry to undergraduates and several graduate level courses in biochemistry. He serves as Assistant Director of SER-CAT at the Advanced Photon Source, Argonne National Laboratory, one of the world's most productive facilities for X-ray structural biology.

Dr. Wendy Dustman is an Assistant Professor in the Department of Microbiology at the University of Georgia, where she teaches Introduction to Microbiology and other undergraduate courses in microbiology. She was a 2009 Lilly Teaching Fellows finalist, and received the 2011 UGA Sandy Beaver Excellence in Teaching Award for the Division of Biological Sciences.

Dr. Julie Kittleson is an Assistant Professor in the Department of Mathematics and Science Education at the University of Georgia. She teaches several undergraduate and graduate level science education classes, including Philosophy and Leadership in Science Classroom Practice. She currently serves on the Board of Reviewers for Science Education.
The UGA Biotech Boot Camp is a one-week (M-F) resident workshop in biotechnology for Georgia high school science teachers aimed at increasing the teacher's content knowledge and skill proficiency in the rapidly growing field of biotechnology. Each day of the Boot Camp consists of morning sessions addressing content knowledge, afternoon sessions focused on laboratory skills, along with lunch and evening activities aimed at improving teaching effectiveness (see biotechbootcamp.uga.edu for the complete schedule).
These afternoon sessions include participation in several hands-on, low or no-cost classroom activities and simulations associated with the above experiments. Computer-based activities, such as the bioinformatics and protein structure-function exercises, are very appealing to the teachers, since schools usually have a computer lab and Internet access. Teachers are shown how to access and use: (1) UniProt Knowledgebase for protein sequences,2 (2) BLAST for sequence comparison,3 (3) the RCSB PDB for 3D structures of proteins and nucleic acids, and (4) PubMed4 and Google Scholar5 for literature searches. Exercises and ideas about how to incorporate these resources into their classroom instruction are provided. For example, in the Classroom Activity Connecting DNA to Disease Using BLAST, a DNA sequence belonging to a disease related protein is given to the student. The student then transcribes and translates the DNA sequence to produce the corresponding amino acid sequence. Using the amino acid sequence, the student then uses BLAST to compare sequences and UniProt to identify and characterize the protein and the disease to which it is associated. Based on this information and using other tools, the student then is asked to describe the protein, its chromosome location and function, the symptoms of the disease, the prognosis and possible treatments.
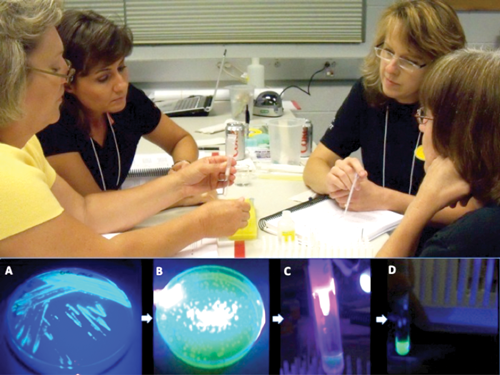
Expression and purification of GFP. (A) Induced cells glowing green under UV light, (B) GFP colonies illuminated by UV, (C) GFP eluting from the ion exchange column illuminated by UV and (D) Purified GFP glowing green under UV light. Teachers: Cindy Kay Plumly, Debbie Thomlinson, Laurie Diffie, Denita Alonso.
The Boot Camp is generally held in early June, when teachers are out of class and beginning to develop lesson plans for the next school year. The workshop hosts between 12-15 high school science teachers and is held at the NESPAL8 facility located on the University of Georgia Tifton campus. The program provides teachers with meals and lodging through the nearby Abraham Baldwin Agricultural College. The workshop provides teachers with (1) a DNA model kit for use in their classroom, (2) a 300 page bound set of workshop (lecture/lab/activity) notes, (3) a USB memory stick containing over 900 Mbytes of workshop related materials (notes, reference papers, experiments and a variety of other classroom activities), (4) Ellyn Daugherty's text book Biotechnology Science for the New Millennium, and (5) a wonderful poster provided by the RCSB PDB.
The workshop has been designed to foster sustained contact between the teachers and workshop personnel via a Science Facilitator who travels to the teacher's classroom and helps the teacher conduct one of the Boot Camp lab activities. Since each teacher attending the workshop represents between 80-130 students each semester, we estimate that over 2500 students per year benefit from skills and knowledge his/her teacher brings back from the workshop. The Boot Camp is funded by an Improving Teacher Quality grant to the University of Georgia School of Education from the U.S. Department of Education. For more information please see www.biotechbootcamp.uga.edu.
- Southeast Regional Collaborative Access Team (SER-CAT) Sector 22 at the Advanced Photon Source, Argonne National Laboratory, www.ser-cat.org.
- UniProt Consortium (2012) Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 40: D71-75.
- S. F. Altschul, W. Gish, W. Miller, E. W. Myers, D. J. Lipman (1990) Basic local alignment search tool. J. Mol. Biol. 215: 403-410 blast.ncbi.nlm.nih.gov.
- E. W. Sayers, T. Barrett, D. A. Benson, E. Bolton, S. H. Bryant, K. Canese, V. Chetvernin, D. M. Church, M. Dicuccio, S. Federhen, M. Feolo, I. M. Fingerman, L. Y. Geer, W. Helmberg, Y. Kapustin, S. Krasnov, D. Landsman, D. J. Lipman, Z. Lu, T. L. Madden, T. Madej, D. R. Maglott, A. Marchler- Bauer, V. Miller, I. Karsch-Mizrachi, J. Ostell, A. Panchenko, L. Phan, K. D. Pruitt, G. D. Schuler, E. Sequeira, S. T. Sherry, M. Shumway, K. Sirotkin, D. Slotta, A. Souvorov, G. Starchenko, T. A. Tatusova, L. Wagner, Y. Wang, W. J. Wilbur, E. Yaschenko, J. Ye. (2012) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40: D13-25. www.ncbi.nlm.nih.gov/pubmed/.
- Butler, D. (2004). Science searches shift up a gear as Google starts Scholar engine. Nature 432(7016): 423. scholar.google.com.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng, T. E. Ferrin. (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605-1612. www.cgl.ucsf.edu/chimera
- Hodis, E., J. Prilusky, et al. (2008). Proteopedia - a scientific 'wiki' bridging the rift between three-dimensional structure and function of biomacromolecules. Genome Biol 9(8): R121. Web site: www.proteopedia.org.
- NESPAL: the National Environmentally Sound Production Agriculture Laboratory, a unit University of Georgia's College of Agricultural and Environmental Sciences. Web site: nespal.cpes.peachnet.edu.

