Data Query, Reporting and Access
Website Statistics
Access statistics for the second quarter of 2012 are shown.
Month |
Unique Visitor |
Visits |
Bandwidth |
July |
184117 |
478089 |
697.83 GB |
August |
175961 |
439848 |
484.91 GB |
September |
241088 |
572574 |
755.14 GB |
Smart Searching with the Top Bar Suggestion Box
Enter text in the top search bar to quickly find structures based on author, macromolecule name, sequence, ligand name or ID, and more.
The top search bar helps users easily and intuitively create simple text searches.
Typing text in the top search bar launches an interactive pop-up box that suggests possible matches that are organized by categories ranging from author name to ontology terms. The top search can be also be limited to quick searches on Author, Macromolecule name, Sequence, or Ligand by selecting the related icon. Using these categories helps to quickly differentiate between possible search results.
For example, autosuggestions for the input bird include authors whose names contain “bird” and structures from the organism bird.
Users who want to perform a simple, non-categorized text search can click the magnifying glass icon or press return.
The top menu bar also recognizes particular types of syntax. Entering SMILES strings will suggest options to perform substructure, exact structure, or similar chemical structure searches, while typing in a sequence will offer different BLAST search options.
Simple text searches complement other RCSB PDB tools: the Explore Archive widget provides browsable data distribution summaries, Browse Database explores the PDB archive using different hierarchical trees, and Advanced Search combines multiple searches of specific types of data.
Different Ways To Explore New Entries
On average, the wwPDB releases 170 structures into the PDB archive each week. The RCSB PDB offers different ways of exploring these new entries:
The Latest Structures widget on the home page provides a slideshow of individual entries. It displays the entry title, image, citation, and a link to the PubMed abstract, if available. Users can pause the show at any point to read the entire abstract, click on the entry title to view the entry's Structure Summary page, or go straight to the Jmol view of the entry.
The set of recently released structures can be launched in the Query Results Browser by clicking on the date listed at the top of the page and the link provided in the New Structures widget on the home page. From the Query Result Browser, users can drill through by category (organism, taxonomy, experimental method, and more), generate reports, and download structure and sequences files.
The MyPDB service can be set to run saved searches with each update. Email alerts (weekly or monthly) will be sent when new entries matching the search are released in the PDB archive.
Download RCSB PDB Mobile
The new app provides fast, on-the-go access. Search the entire PDB, view the latest weekly release of structures, access your MyPDB account, view the entire catalog of Molecule of the Month articles, and more using either a WiFi or cellular data connection and an iPad/iPhone device.
The latest release improves upon the 3D display of macromolecular biomolecules.
A version of the app for the Android platform is in development.

Search the PDB, access MyPDB, view molecules in 3D, and more with RCSB PDB Mobile.
Build Complex Queries with Advanced Search
 Advanced Search provides the capability of combining multiple searches of specific types of data in a logical AND or OR. The result is a list of structures that comply with ALL or ANY of the search criteria, respectively.
Advanced Search provides the capability of combining multiple searches of specific types of data in a logical AND or OR. The result is a list of structures that comply with ALL or ANY of the search criteria, respectively.
Individual data items are organized by category; contextual help and examples are available by selecting the question mark icon. New options include quick searches by experimental and/or molecule type, searches based on structure determination, and the ability to find structures containing interresidue connectivity (LINK records) that cannot be inferred from the primary structure.
Currently, users can build searches based on:
- Quick Search: retrieve all PDB entries or a subset based on experimental method or molecule type
- ID(s) and Keywords: PDB, PubMed, UniProtKB, Pfam IDs; text and keyword searching
- Structure Annotation: structure title, description; and macromolecule name
- Deposition: author name; deposit, release, and revision date; latest released and modified structures; Structural Genomics Project
- Structure Features: macromolecule type; number of chains (asymmetric unit or biological assembly), entities, models, and disulfide bonds; interresidue connectivity (LINK records); molecular weight; secondary structure content; secondary structure length; SCOP, CATH, taxonomy
- Sequence Features: sequence; translated nucleotide sequence; sequence motif; chain length; genome location
- Chemical Components: name; ID; InChi descriptor; SMILES/SMARTS; molecular weight; chemical formula; chemical component type; binding affinity; has ligands; has modified residues; sub-components
- Biology: Source; expression organism; Enzyme Classification; biological process; cell component; molecular function; Transporter Classification
- Methods: experimental method; X-ray resolution, R factor, diffraction source, structure determination method, reflections, cell dimensions, software, space group, crystal properties, detector; EM assembly
- Publication: citation; MeSH terms; PubMed abstract
- Misc: Has external links
Advanced Searches can be stored in MyPDB to be run or modified at any time.
Customized Home Page: Structure Comparison Tool
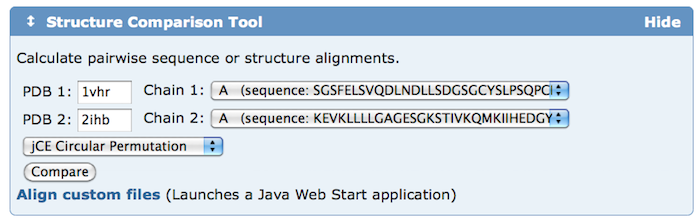 Add the Structure Comparison Tool widget to your home page to quickly calculate pairwise sequence and structure alignments.
Add the Structure Comparison Tool widget to your home page to quickly calculate pairwise sequence and structure alignments.
The RCSB PDB homepage is comprised of web widgets that can be moved around, minimized, or hidden so users can create a website that reflects their interests. Frequently used features can be moved to the top, while less popular items can be hidden or collapsed.
The Structure Comparison Tool widget can be added so it appears on every visit to the RCSB PDB home page. The tool calculates pairwise sequence (blast2seq, Needleman-Wunsch, and Smith-Waterman) and structure alignments (FATCAT, CE, Mammoth, TM-Align, TopMatch).
Comparisons can be made for any protein in the PDB archive and for customized or local files not in the PDB. Special features include support for both rigid-body and flexible alignments (via jFATCAT) and detection of circular permutations (via jCE).
To add this widget to the RCSB PDB home page, select the Customize This Page button from the left menu. Download Files, Sequence Search, and ADIT Deposition widgets can also be added or removed.
The Comparison Tool is also available from Structure Summary pages and as a stand-alone Java Web Start application, with additional information available online.
Explore Different PDB Data Overviews
 The RCSB PDB maintains several lists that display PDB data in interesting and useful ways. Highlights include:
The RCSB PDB maintains several lists that display PDB data in interesting and useful ways. Highlights include:
- Proteins Solved by Multiple Experimental Methods: Lists clusters of proteins (with greater than 95% sequence similarity) containing at least one structure solved by one method (e.g. X-ray) and one by a different method (e.g. NMR).
- PDB Statistics: Content Distribution & Growth: View data distributions of the PDB archive based on characteristics like space group, journal, EC, and more. The growth of the number of PDB entries released per year is organized around experimental method, molecule type, and unique protein classifications. Data can be downloaded as Excel documents.
- RSS Feeds: Subscribe to RSS feeds to access the latest structures (from the RCSB PDB) and Molecule of the Month articles (from PDB-101) as soon as they become available.
- Summaries of PDB Data: Links to summary files available from the PDB FTP, including a file with all PDB sequences in FASTA format; one with all PDB IDs, molecule type, and experimental method; and one with all PDB IDs and authors.
- Secondary Structure Files: A FASTA formatted file generated using DSSP displays sequences and secondary structure for all entries. A separate file includes notation of regions which have not been experimentally observed in addition to the secondary structure.
- Latest weekly release and all released entries: The top right of every RCSB PDB page header links to the full archive (select the number of total structures) and to the entries in the most recent update (select the date listed).
- PDB-101 Author Profiles: Author Profiles vertically display structures associated with a particular researcher. Profiles are also available for Structural Genomics centers.
- PDB-101 Molecule of the Month: Articles are indexed by title, date, and category, and accessible from an alphabetical pull-down menu.
Improved Ligand Reports
Ligand Summary Reports include information about the selected chemical components such as formula, molecular weight, name, SMILES string, which PDB entries are related to the ligand, and how they are related.
All tabular report features are also available, including sorting, filtering, export to other report formats, and column customization.
Ligands can also be displayed as an image collage.
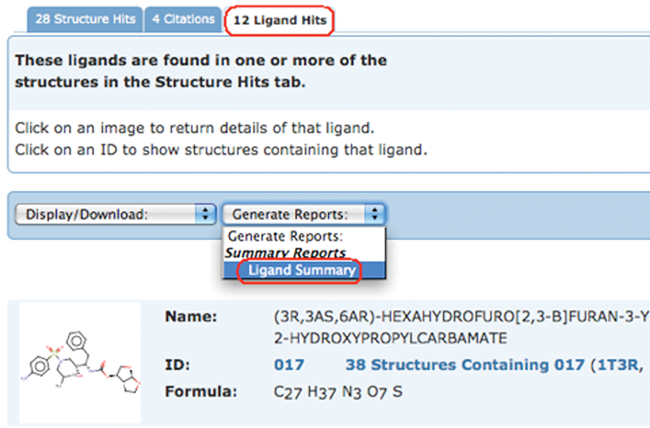
To generate a report, select the Ligand Summary option from the Generate Reports pull-down menu for any set of query results.
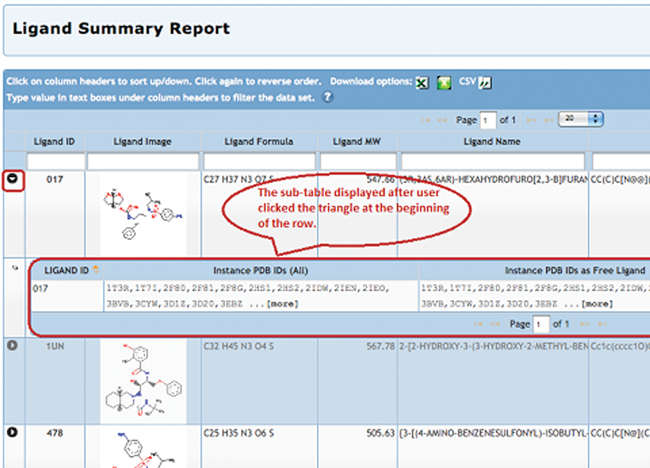
For each ligand included in the report, a sub-table can be selected to show lists of all related PDB entries that contain the ligand, the entries that contain the ligand as a free ligand, and entries that
contain the ligand as part of a larger, polymeric ligand.
To display this sub-table, select the triangle shown next to the Ligand ID. The display is limited to 15 PDB IDs in each column. For ligands associated with more than 15 PDB entries, the ... [more] link will launch a query for that set of structures.